1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008; 58:71–96. PMID:
18287387.

2. Hu WP, Yu HS, Chen YR, Tsai YM, Chen YK, Liao CC, Chang LS, Wang JJ. Synthesis and biological evaluation of thiobenzanilides as anticancer agents. Bioorg Med Chem. 2008; 16:5295–5302. PMID:
18359635.

3. Kemnitzer W, Jiang S, Wang Y, Kasibhatla S, Crogan-Grundy C, Bubenik M, Labrecque D, Denis R, Lamothe S, Attardo G, Gourdeau H, Tseng B, Drewe J, Cai SX. Discovery of 4-aryl-4H-chromenes as a new series of apoptosis inducers using a cell- and caspase-based HTS assay. Part 5: modifications of the 2- and 3-positions. Bioorg Med Chem Lett. 2008; 18:603–607. PMID:
18077161.

4. Wordenmam L, Mitchison TJ. Dynamics of microtubule assembly in vivo. Mod Cell Biol. 1994; 13:287–301.
5. Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004; 4:253–265. PMID:
15057285.

6. Xia Y, Yang ZY, Xia P, Bastow KF, Tachibana Y, Kuo SC, Hamel E, Hackl T, Lee KH. Antitumor agents. 181. Synthesis and biological evaluation of 6,7,2',3',4'-substituted-1,2,3,4-tetrahydro-2-phenyl-4-quinolones as a new class of antimitotic antitumor agents. J Med Chem. 1998; 41:1155–1162. PMID:
9544215.

7. Rowinsky EK, Donehower RC. The clinical pharmacology and use of antimicrotubule agents in cancer chemotherapeutics. Pharmacol Ther. 1991; 52:35–84. PMID:
1687171.

8. Verweij J, Clavel M, Chevalier B. Paclitaxel (Taxol) and docetaxel (Taxotere): not simply two of a kind. Ann Oncol. 1994; 5:495–505. PMID:
7918121.
9. Shi Q, Chen K, Li L, Chang JJ, Autry C, Kozuka M, Konoshima T, Estes JR, Lin CM, Hamel E. Antitumor agents, 154. Cytotoxic and antimitotic flavonols from Polanisia dodecandra. J Nat Prod. 1995; 58:475–482. PMID:
7623025.

10. Manthey JA, Grohmann K, Guthrie N. Biological properties of citrus flavonoids pertaining to cancer and inflammation. Curr Med Chem. 2001; 8:135–153. PMID:
11172671.

11. Hodek P, Trefil P, Stiborová M. Flavonoids-potent and versatile biologically active compounds interacting with cytochromes P450. Chem Biol Interact. 2002; 139:1–21. PMID:
11803026.

12. Manthey JA, Guthrie N. Antiproliferative activities of citrus flavonoids against six human cancer cell lines. J Agric Food Chem. 2002; 50:5837–5843. PMID:
12358447.

13. Chan HY, Wang H, Leung LK. The red clover (Trifolium pratense) isoflavone biochanin A modulates the biotransformation pathways of 7,12-dimethylbenz[a]anthracene. Br J Nutr. 2003; 90:87–92. PMID:
12844379.
14. Lee D, Bhat KP, Fong HH, Farnsworth NR, Pezzuto JM, Kinghorn AD. Aromatase inhibitors from Broussonetia papyrifera. J Nat Prod. 2001; 64:1286–1293. PMID:
11678652.

15. Pouget C, Fagnere C, Basly JP, Besson AE, Champavier Y, Habrioux G, Chulia AJ. Synthesis and aromatase inhibitory activity of flavanones. Pharm Res. 2002; 19:286–291. PMID:
11934235.
16. Lai YY, Huang LJ, Lee KH, Xiao Z, Bastow KF, Yamori T, Kuo SC. Synthesis and biological relationships of 3',6-substituted 2-phenyl-4-quinolone-3-carboxylic acid derivatives as antimitotic agents. Bioorg Med Chem. 2005; 13:265–275. PMID:
15582470.

17. Li L, Wang HK, Kuo SC, Wu TS, Lednicer D, Lin CM, Hamel E, Lee KH. Antitumor agents. 150. 2',3',4',5',5,6,7-substituted 2-phenyl-4-quinolones and related compounds: their synthesis, cytotoxicity, and inhibition of tubulin polymerization. J Med Chem. 1994; 37:1126–1135. PMID:
8164254.

18. Zhang SX, Bastow KF, Tachibana Y, Kuo SC, Hamel E, Mauger A, Narayanan VL, Lee KH. Antitumor agents. 196. Substituted 2-thienyl-1,8-naphthyridin-4-ones: their synthesis, cytotoxicity, and inhibition of tubulin polymerization. J Med Chem. 1999; 42:4081–4087. PMID:
10514278.

19. Chen K, Kuo SC, Hsieh MC, Mauger A, Lin CM, Hamel E, Lee KH. Antitumor agents. 174. 2',3',4',5,6,7-Substituted 2-phenyl-1,8-naphthyridin-4-ones: their synthesis, cytotoxicity, and inhibition of tubulin polymerization. J Med Chem. 1997; 40:2266–2275. PMID:
9216846.

20. Litvinov VP. Chemistry and biological activities of 1,8-naphthyridines. Russ Chem Rev. 2004; 73:637–670.

21. Kren V, Rezanka T. Sweet antibiotics - the role of glycosidic residues in antibiotic and antitumor activity and their randomization. FEMS Microbiol Rev. 2008; 32:858–889. PMID:
18647177.

22. Tsuzuki Y, Tomita K, Shibamori K, Sato Y, Kashimoto S, Chiba K. Synthesis and structure-activity relationships of novel 7-substituted 1,4-dihydro-4-oxo-1-(2-thiazolyl)-1,8-naphthyridine-3-carboxylic acids as antitumor agents. Part 2. J Med Chem. 2004; 47:2097–2109. PMID:
15056007.

23. Tomita K, Tsuzuki Y, Shibamori K, Tashima M, Kajikawa F, Sato Y, Kashimoto S, Chiba K, Hino K. Synthesis and structure-activity relationships of novel 7-substituted 1,4-dihydro-4-oxo-1-(2-thiazolyl)-1,8-naphthyridine-3-carboxylic acids as antitumor agents. Part 1. J Med Chem. 2002; 45:5564–5575. PMID:
12459024.

24. Srivastava SK, Jha A, Agarwal SK, Mukherjee R, Burman AC. Synthesis and structure-activity relationships of potent antitumor active quinoline and naphthyridine derivatives. Anticancer Agents Med Chem. 2007; 7:685–709. PMID:
18045063.

25. Tsuzuki Y, Tomita K, Sato Y, Kashimoto S, Chiba K. Synthesis and structure-activity relationships of 3-substituted 1,4-dihydro-4-oxo-1-(2-thiazolyl)-1,8-naphthyridines as novel antitumor agents. Bioorg Med Chem Lett. 2004; 14:3189–3193. PMID:
15149673.

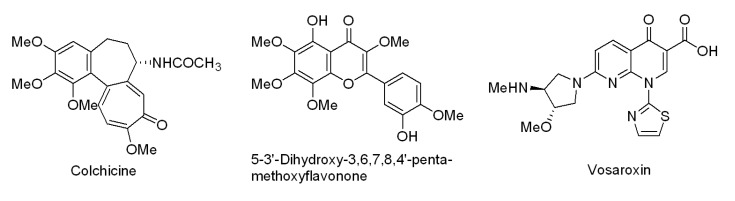
26. Abbas JA, Stuart RK. Vosaroxin: a novel antineoplastic quinolone. Expert Opin Investig Drugs. 2012; 21:1223–1233.

27. Deady LW, Rogers ML, Zhuang L, Baguley BC, Denny WA. Synthesis and cytotoxic activity of carboxamide derivatives of benzo[b][1,6]naphthyridin-(5H)ones. Bioorg Med Chem. 2005; 13:1341–1355. PMID:
15670942.

28. Park JG, Kramer BS, Steinberg SM, Carmichael J, Collins JM, Minna JD, Gazdar AF. Chemosensitivity testing of human colorectal carcinoma cell lines using a tetrazolium-based colorimetric assay. Cancer Res. 1987; 47:5875–5879. PMID:
3664487.
29. Manthey JA, Guthrie N. Antiproliferative activities of citrus flavonoids against six human cancer cell lines. J Agric Food Chem. 2002; 50:5837–5843. PMID:
12358447.

30. Hwang YJ, Park SM, Yim CB, Im C. Cytotoxic activity and quantitative structure activity relationships of arylpropyl sulfonamides. Korean J Physiol Pharmacol. 2013; 17:237–243. PMID:
23776401.

31. Lim JC, Park SY, Nam Y, Nguyen TT, Sohn UD. The protective effect of eupatilin against hydrogen peroxide-induced injury involving 5-lipoxygenase in feline esophageal epithelial cells. Korean J Physiol Pharmacol. 2012; 16:313–320. PMID:
23118554.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download