Abstract
Figures and Tables
Fig. 1

Fig. 2
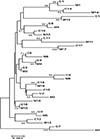
Fig. 3
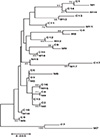
Fig. 4

Journal List > J Korean Med Sci > v.29(4) > 1022526




Hyoung Su Kim 
https://orcid.org/http://orcid.org/0000-0002-2394-1095
Bo Youn Choi 
https://orcid.org/http://orcid.org/0000-0002-5661-9879
Hyeok Soo Choi 
https://orcid.org/http://orcid.org/0000-0003-0924-5912
Woon Geon Shin 
https://orcid.org/http://orcid.org/0000-0002-9851-5576
Kyung Ho Kim 
https://orcid.org/http://orcid.org/0000-0002-9607-6258
Jin Heon Lee 
https://orcid.org/http://orcid.org/0000-0003-3561-3870
Hak Yang Kim 
https://orcid.org/http://orcid.org/0000-0002-7459-3473
Myoung Kuk Jang 
https://orcid.org/http://orcid.org/0000-0001-7968-7543
Dong Joon Kim 
https://orcid.org/http://orcid.org/0000-0002-5792-1500
Myung Seok Lee 
https://orcid.org/http://orcid.org/0000-0001-7237-7761
Choong Kee Park 
https://orcid.org/http://orcid.org/0000-0003-1947-345X