Abstract
Two variants of the inosine triphosphatase (ITPA: rs1127354, rs7270101) gene cause ITPA deficiency and protect against the hemolytic toxicity of ribavirin. We investigated the clinical significance of ITPA variants in Korean patients treated with pegylated interferon (PEG-IFN) plus ribavirin. Of the 133 patients, 108 were CC and 25 were non-CC at rs1127354 (groups A and B, respectively). On the other hand, at rs7270101 all 133 were AA. The mean values of Hemoglobin (Hgb) after 4, 8, and 12 weeks of treatment in groups A and B were 12.2 and 14.0, 11.8 and 13.2, and 11.5 and 12.9, respectively (P=0.001, 0.036, 0.036). Sustained virologic response (SVR) was achieved in 67.8% (40/59) of genotype 1 patients and in 75% (27/36) of non-genotype 1 patients. Regarding ITPA variants, SVR was achieved by 66% and 80% of genotype 1 (P=0.282), and by 78% and 71% (P=0.726) of non-genotype 1. SVR was not significantly different in groups A and B. In conclusion, non-CC at rs1127354 without involvement of rs7270101 is strongly associated with protection from ribavirin-induced anemia, however, ITPA genotype is not associated with SVR.
Hepatitis C virus (HCV) infection affects approximately 170 million people worldwide, and 3-4 million individuals are newly infected each year (1, 2). Furthermore, hepatitis C infection is a major cause of serious liver diseases, such as, liver cirrhosis and hepatocellular carcinoma (3). The standard anti-viral treatment for chronic hepatitis C (CHC) patient is combination of pegylated interferon (PEG-IFN) and ribavirin (RBV), and the purpose of anti-viral treatment is sustained virologic response (SVR), defined as negativity for HCV RNA for 24 weeks after treatment cessation (4, 5). However, the side effects of treatment present significant problems. In particular, RBV-induced anemia is one of the most important side effects, and up to 15% of patients experience dose modification. Accordingly, optimal treatment may require individualized approaches in CHC patients (5).
RBV is a synthetic guanosine analogue and a prodrug, which resembles purine RNA nucleotides after undergoing metabolism, and interferes with the RNA metabolism required for viral replication (6). RBV is directly toxic to erythrocytes and is associated with hemolysis via guanosine triphosphate pool depletion and the acceleration of erythrocytosis. However, RBV-induced anemia is usually reversible and dose-related (7, 8).
A new era was brought into being by the completion of the Human Genome Project, which included the genome-wide association study (GWAS). According to several recent studies, genetic polymorphisms located near the interleukin 28B (IL28B) gene affect virologic response to treatment (9). In another study, polymorphisms of the inosine triphosphatase (ITPA) gene in chromosome 20 were found to influence RBV-induced anemia in CHC patients, and confirmed the presence of rs1127354 (a missense variants in exon 2) and rs7270101 (a splice-altering single nucleotide polymorphism [SNP] located in the second intron) (10).
However, most research on this topic has been carried out in the USA and Europe. In this study, the two main ITPA variants, which have been shown to be associated with RBV-induced anemia (rs1127354 and rs7270101) were investigated in Korean CHC patients treated with PEG-IFN plus RBV. In addition, we investigated the IL28B SNP, which has been shown to be most strongly associated with virologic response in genotype 1 patients.
In this retrospective cohort study, we studied 133 Korean patients with CHC infection treated at Gachon University Gil Medical Center from January 2008 to December 2011. Inclusion criteria were: 1) a diagnosis of CHC infection; 2) HCV genotype confirmed using the core region of HCV cDNA by polymerase chain reaction (PCR) and subsequent DNA sequencing; 3) a HCV RNA level of ≥1,000 IU/mL determined by PCR using an Abbott m2000rt instrument (Abbott Laboratories, Chicago, IL) with a serum HCV RNA detection limit of <50 IU/mL; and 4) Korean ethnicity and an age from 20 to 70 yr. Exclusion criteria were: 1) decompensated liver cirrhosis; 2) hepatitis B surface antigen positivity; 3) the presence or a history of hepatocellular carcinoma or 4) of other liver diseases, such as, autoimmune hepatitis, alcoholic liver disease, or a chronic liver disease other than CHC; 5) chronic renal disease, or 6) anemia before treatment (hemoglobin ≤12 g/dL), absolute neutrophil count ≤500 or thrombocytopenia (platelets ≤100,000/µL).
All patients were treated using a standard PEG-IFN alfa-2a plus RBV therapy. Patients with HCV genotype 1 were injected subcutaneously with 180 µg of PEG-IFN alpha-2a (Pegasys®, F. Hoffmann-La Roche, Ltd., Basel, Switzerland) per week regardless of weight plus 1,200 mg of RBV daily for patients >75 kg or 1,000 mg daily for patients <75 kg. Patients with HCV genotype 2 or 3 were treated with 180 µg of PEG-IFN alfa-2a s.c. weekly and 800 mg of RBV daily. This standard combination therapy was continued for 48 weeks in patients with genotype 1 and for 24 weeks in patients with genotype 2 or 3.
The following were documented before anti-viral treatment: age, gender, sex, body mass index (BMI), and biochemical parameters, hemoglobin (Hgb), white blood cells, neutrophils, platelets, alanine transaminase (ALT) level, HCV genotype, and HCV RNA level. In addition, we checked Hgb level and RBV dose before and after 4, 8, and 12 weeks of anti-viral treatment, and checked serum HCV RNA levels before and after 4 and 12 weeks of anti-viral treatment, and at 24 weeks after terminating anti-viral treatment to investigate rapid virologic response (RVR), early virologic response (EVR), and sustained virologic response (SVR).
Blood was collected into EDTA tubes, and genomic DNA was extracted from whole blood using the QIAamp® DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA qualities were assessed by calculating absorbance ratios (OD 260 nm/280 nm) using a NanoDrop model ND-1000 (Thermo Fisher Scientific Inc., Wilmington, DE, USA) and DNA quantities were estimated using the Quibit® dsDNA BR Assay kit using a Quibit® Fluorometer (Invitrogen, Carlsbad, CA, USA).
ITPA variants rs1127354 C>A, rs7270101 A>C were determined in whole blood samples using the validated Pyrosequencing™ assay. Primers were designed using PQS Assay Design software (Qiagen, Hilden, Germany) to target two SNP regions. The primers used were rs1127354: 5'-biotin-CGTGCTCACATGGAGAATCA-3' (forward primer), 5'-TTTTCTGTGCCACCAAAGTG-3' (reverse primer), and rs7270101: 5'-TTGGTGGCACAGAAAATTGAC-3' (forward primer), 5'-biotin-GGGAAACAGACACACAGAAAGTCA-3' (reverse primer). PCR reactions were carried out by adding 20 ng of template DNA, 5 µL of 10× PCR buffer, 5 µL of 2.5 mM dNTPs, 1 µL of each 10 µM forward and reverse primer (one primer was biotinylated), and 0.5 µL of Blend Taq plus DNA polymerase (Toyobo, Osaka, Japan) in a 50 µL reaction mix in a 96-well plate. Amplification was performed under the following conditions: rs1127354-initial denaturation at 94℃ for 2 min followed by 45 cycles of 94℃ for 30 sec, 60℃ for 30 sec, 72℃ for 30 sec; rs7270101-initial denaturation at 94℃ for 2 min followed by 45 cycles of 94℃ for 30 sec, 58℃ for 30 sec, and 72℃ for 30 sec.
IL28B variant rs8099917 T>G was identified in whole blood using validated Pyrosequencing™ assays. The primers used were rs8099917: 5'-biotin-TCCTCCTTTTGTTTTCCTTTCTG-3' (forward primer), 5'-AAAAAGCCAGCTACCAAACTGT-3' (reverse primer). Amplification was performed under the following conditions: rs8099917-initial denaturation at 94℃ for 2 min followed by 45 cycles of 94℃ for 30 sec, 60℃ for 30 sec, and 72℃ for 30 sec.
Pyrosequencing was performed took using an automated PSQ 96 MA instrument and the PyroMark Gold Q96 reagent kit for SNP genotyping and mutation analysis (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The sequencing primers used to detect short DNA sequences around SNPs of interest were: rs8099917-5'-TTCCAATTTGGGTGA-3', rs1127354-5'-TTCAGATTCTAGGAGATAAGTT-3', and rs7270101-5'-GAAATCCAACCATCTTTTA-3'.
The primary end-points of this study were Hgb decline and rate of RBV dose reduction after 4, 8, and 12 weeks of treatment, and the secondary end-point was virologic response. SVR was defined as an undetectable serum HCV RNA level by real-time PCR assay at 24 weeks after termination of the anti-viral treatment. RVR was defined as undetectable serum HCV RNA level after 4 weeks of treatment, and EVR as an undetectable or a ≥2 log drop in serum HCV RNA after 12 weeks of treatment.
Student t-test and chi-square test were used to estimate and compare significant Hgb declines after 4, 8, and 12 weeks of treatment. Associations between ITPA, IL28B variants, and virologic response (RVR, EVR, and SVR) were examined using the chi-square test. The potential predictors of a decrease in hemoglobin subjected to univariate analysis were; gender, age, BMI, liver cirrhosis, hemoglobin level, and ITPA variants, and the potential predictors of SVR subjected to univariate analysis were; gender, age, BMI, liver cirrhosis, genotype, RVR, EVR, and RBV dose reduction, ITPA, and IL28B variant. Multivariate analysis included all predictors identified by univariate analysis was performed by multiple logistic regression analysis. In addition, odds ratios (ORs) with 95% confidence interval (CI) were determined. Statistical significance was accepted for P values<0.05, and the analysis was conducted using SPSS II v. 12 software (SPSS, Chicago, IL, USA).
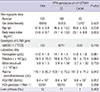
One hundred and thirty three patients diagnosed as CHC were treated with PEG-IFN-α-2b and RBV from 1, January 2008 to 31, December 2011 at the gastroenterology department of Gil Medical Center. Of the 133 patients, 82 (61%) were HCV genotype 1 and 51 (39%) were HCV non-genotype 1. Proportions of HCV non-genotype 1 were; HCV genotype 2 (n=49, 96%) and HCV genotype 3 (n=2, 4%). We investigated baseline genetic distribution and clinical characteristics with respect to the ITPA polymorphisms (rs1127354 and rs7270101) which related to RBV induced anemia. For rs1127354, there were 108 patients of the CC genotype and 25 patients of the CA/AA genotype, and for rs7270101, all 133 patients were of the AA genotype (Fig. 1). Baseline characteristics of the 108 CC patients and 25 CA/AA patients (groups A and B) are summarized in Table 1. No significant intergroup differences were found for sex, age, IL28B gene, initial Hgb, platelet count, white blood cell count, serum HCV RNA, HCV genotype, and liver cirrhosis. However, mean BMI and mean alanine aminotransferase level were higher in group B (P=0.042 and P=0.033, respectively).
We investigated time dependent Hgb declines between groups A and B after anti-viral treatment. RBV induced anemia was defined as a decline of more than 3 g/dL of Hgb level from before anti-viral treatment or less than 10 g/dL of Hgb level during anti-viral treatment (10, 11). Twenty-one (19%) patients in group A and 1 (4%) patient in group B were diagnosed with RBV induced anemia after 4 weeks of anti-viral treatment, 45 (42%) and 2 (8%) after 8 weeks, and 54 (50%) and 4 (16%) after 12 weeks. Mean Hgb levels were 12.2, 11.8, and 11.5 g/dL after 4, 8, and 12 weeks of antiviral treatments in group A and 14, 13.2, and 12.9 g/dL in group B (P=0.001, 0.036, 0.036). Fig. 2 compares decreases in mean Hgb values in the two groups. Additionally, we conducted subgroup analysis between group A and B of HCV genotype 1. Mean Hgb levels were 12.4, 11.7, and 11.3 after 4, 8 and 12 weeks of antiviral treatment in group A of HCV genotype 1 and 13.9, 13.3, and 12.7 in group B (P=0.206, 0.601, 0.155).
RBV dose modifications were as followings; a reduction of 200 mg when the Hgb level was<10 g/dL and stopped when the Hgb level was<8 g/dL. Eight patients discontinued PEG-IFN and RBV treatment because of side effects, such as, a loss of appetite, dizziness, or depression, and therefore, the analysis of RBV dose modification included 125 patients. During the first 12 weeks of anti viral treatment, RBV dose reduction (>200 mg) was performed in 40 patients. RBV dose reduction was started earlier in 16 (16%) patients in group A than in the 3 (14%) patients in group B. During the first 12 weeks of anti-viral treatment, the rate of RBV dose reduction increased steadily in group A, whereas no further RBV dose reduction occurred after 4 weeks in group B. RBV dose reductions after 12 weeks of treatment were performed in 37 patients (36%) in group A and in 3 patients (14%) in group B (P=0.042) (Fig. 3).
To investigate the influences of potential prognostic factors on RBV induced anemia, 6 factors (gender, age>60 yr, BMI>23 kg/m2, liver cirrhosis, initial Hgb level [<14 g/dL], and CC genotype of the ITPA variant) were examined individually by univariate analysis. To identify independent prognostic factors, stepwise forward multiple logistic regression analysis was performed. Multivariate analysis showed that only a male gender and the CC genotype positively influenced RBV induced anemia (Table 2).
SVR was achieved in 67.8% (40/59) of genotype 1 patients and in 75% (27/36) of non-genotype 1 patients. Regarding ITPA variants, SVR was achieved by 66% and 80% of genotype 1 (P=0.282), and by 78% and 71% (P=0.726) of non-genotype 1. SVR was not significantly different in both groups (Fig. 4).
To determine whether ITPA variant affects virological response, logistic regression analysis was used. A total of 125 patients who completed the anti-viral treatment were analyzed. Initially 10 factors, that is, gender, age (<60 yr), BMI>23 kg/m2, liver cirrhosis, genotype 1, RVR, EVR, RBV dose reduction (<20%), CC genotype of ITPA variant, and the recently reported IL28B variant (rs8099917, TT), were examined individually by univariate analysis. Stepwise forward multiple logistic regression analysis was performed to identify independent factors. ITPA variant (rs1127354) showed no significant association (P=0.907). However, age, EVR, and IL28B (rs8099917, TT) remained significant by multivariate analysis (Table 3).
Most HCV infections progress to chronic disease and if left untreated and can lead to liver cirrhosis and hepatocellular carcinoma (12, 27). Interferon-alfa monotherapy was first used to treat CHC patients, but the SVR rate achieved was only 15 to 20%. Because of this serious shortcoming, combination therapy with anti-viral or anti-inflammatory drugs was examined. It was found that a combination of PEG-IFN and RBV was most effective, and that it achieved SVR in more than 50% of patients (7). Currently, the standard anti-viral treatment for CHC is a combination of PEG-IFN and RBV (13).
RBV (1-b-D-ribofuranosyl-1H-1,2,4-triazole-3-carboxamide) is a synthetic nucleoside analogue. Although RBV monotherapy has little antiviral activity against HCV, it improves treatment response and when used in combination with IFN increases the SVR rate to over 56% (14, 15). However, RBV often causes reversible hemolytic anemia, which often makes treatment intolerable (16). The standard dose of RBV used is 1,000-1,200 mg/day, and at this level over 50% of patients experienced a decline in Hgb level (17). Anemia begins early after treatment initiation, and is most serious after 4 weeks of treatment. Associated symptoms, such as, fatigue, decreased quality of life, can also occur. Furthermore, anemia often necessitates treatment withdrawal or RBV dose reduction (18). In addition, some have reported that lowering the RBV dosage may reduce the chance of SVR and increase the rate of relapse (17, 19). The mechanism of RBV induced anemia is still unknown (17), but it has been hypothesized that an accumulation of the active form of RBV in red blood cells inhibits intracellular energy metabolism, causes oxidative membrane damage, and accelerates the extravascular removal of erythrocytes (17, 20).
It is important we can predict RBV induced anemia, and various predictive factors have been proposed. However, it is still difficult to predict the risk of hemolysis before the administration of RBV (21). Recently, several related studies have been conducted. In particular, the GWAS study on HCV infection identified two host genetic SNPs; one in the IL28B gene and the other is the ITPA gene. The former was found to be strongly associated with response to treatment for chronic genotype 1 HCV infections, whereas the latter was found to predict RBV induced anemia (10).
ITPA gene encodes a protein that cleaves inosine triphosphate (ITP). However, the precise cellular function of ITPA has not been elucidated (22). As mentioned above, the GWAS study identified two genetic variants in chromosome 20, namely, rs1127354 (a missense variant in exon2) and rs7270101 (a splice altering SNP). According to the study, these variants are strongly and independently associated with a reduction in Hgb during early PEG-IFN plus RBV treatment in CHC (10, 11).
In our study, a functional SNP in ITPA, rs1127354, was found to be strongly associated with RBV induced anemia among 133 Korean patients (Fig. 2). Of these 133 patients, 108 possessed the RBV-sensitive CC genotype and 25 the RBV-resistant CA/AA genotype, which concur with the results of Western studies (10). However, in contrast to western studies, all Koreans enrolled were monoallelic at rs7270101 and possessed the AA genotype, which is similar to that found in Japan (23, 24). According to previous studies, polymorphisms of the ITPA gene were associated with RBV-induced anemia in HCV genotype 1 (10). In our study, however, ITPA variant was not associated with time dependent Hgb decline in HCV genotype 1. The main cause of this result was probably because single center study conducted in small number of patients.
In our multivariate analysis of anemia after 12 weeks of treatment, gender and rs1127354 were found to be independently associated with RBV induced anemia (Table 3), which suggested that rs1127354 might be a useful predictive marker of RBV induced anemia. According to previous studies, RBV dose reduction due to anemia in patients treated with PEG-IFN plus RBV is influenced by ITPA variants (11). In the present study, RBV dose was reduced more in group A than in group B during first 12 weeks of treatment (36% vs 14%, P=0.042).
Remarkably, despite its protective effect against anemia and less need for RBV dose reduction, multivariate analysis revealed that rs1127354 was not significantly associated with virological response. It is widely recognized that age and IL28B (rs8099917) are associated with SVR (Table 3), and in patients with anemia, it can be inferred that RBV dose reduction could decrease treatment efficacy. However, according to previous studies, the reason of this discrepancy between RBV dose reduction and SVR is controversial (8, 11). There are several possible explanations for this discrepancy. First, other risk factors, such as, an advanced age and histologic disease status, can confuse attempts to determine whether RBV dose reduction is responsible (8). Second, RBV dose reduction is the only mechanism whereby ITPA variants might associated with virological response (11). Third, RBV dose reduction may not be associated with virological response if RBV is not discontinued (25, 26).
This study has limitations that should be mentioned. First, it is limited by its retrospective, single center design. Second, it was conducted in a Korean population, and thus, the results obtained may not be applicable to other ethnicities.
In conclusion, there are strong points in our study. First, ITPA genotype distributions in Koreans and Caucasians differ. Second, non-CC at rs1127354 (regardless of rs7270101 genotype) is strongly associated with protection from RBV induced anemia. Third, RBV dose reduction due to RBV induced anemia is influenced by ITPA variants, and finally, ITPA genotype is not associated with virological response.
Figures and Tables
Fig. 3
Cumulative percentages of patients requiring ribavirin dose reduction after 4, 8, and 12 weeks of treatment by ITPA genotype.

Fig. 4
Virologic response by inosine triphosphatase genotype. RVR, Rapid virologic response; EVR, Early virologic response; SVR, Sustained virologic response.

References
1. Davis GL, Alter MJ, El-Serag H, Poynard T, Jennings LW. Aging of hepatitis C virus (HCV)-infected persons in the United States: a multiple cohort model of HCV prevalence and disease progression. Gastroenterology. 2010; 138:513–521.
2. Wiese M, Grüngreiff K, Güthoff W, Lafrenz M, Oesen U, Porst H. East German Hepatitis C Study Group. Outcome in a hepatitis C (genotype 1b) single source outbreak in Germany: a 25-year multicenter study. J Hepatol. 2005; 43:590–598.
3. Massard J, Ratziu V, Thabut D, Moussalli J, Lebray P, Benhamou Y, Poynard T. Natural history and predictors of disease severity in chronic hepatitis C. J Hepatol. 2006; 44:S19–S24.
4. Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL Jr, Häussinger D, Diago M, Carosi G, Dhumeaux D, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002; 347:975–982.
5. Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001; 358:958–965.
6. Homma M, Hosono H, Hasegawa Y, Kohda Y. Morphological transformation and phosphatidylserine exposure in erythrocytes treated with ribavirin. Biol Pharm Bull. 2009; 32:1940–1942.
7. Sakamoto N, Tanaka Y, Nakagawa M, Yatsuhashi H, Nishiguchi S, Enomoto N, Azuma S, Nishimura-Sakurai Y, Kakinuma S, Nishida N, et al. ITPA gene variant protects against anemia induced by pegylated interferon-α and ribavirin therapy for Japanese patients with chronic hepatitis C. Hepatol Res. 2010; 40:1063–1071.
8. You BC, Kim YS, Kim Hi, Kim SH, Park SS, Seo YR, Kim SG, Lee SW, Kim HS, Jeong SW, et al. A reduced dose of ribavirin does not influence the virologic response during pegylated interferon alpha-2b and ribavirin combination therapy in patients with genotype 1 chronic hepatitis C. Clin Mol Hepatol. 2012; 18:272–278.
9. Jeong SH, Jung YK, Yang JW, Park SJ, Kim JW, Kwon OS, Kim YS, Choi DJ, Kim JH. Efficacy of peginterferon and ribavirin is associated with the IL28B gene in Korean patients with chronic hepatitis C. Clin Mol Hepatol. 2012; 18:360–367.
10. Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, Little LD, Qiu P, Bertelsen AH, Watson M, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010; 464:405–408.
11. Thompson AJ, Fellay J, Patel K, Tillmann HL, Naggie S, Ge D, Urban TJ, Shianna KV, Muir AJ, Fried MW, et al. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010; 139:1181–1189.
12. Lavanchy D. The global burden of hepatitis C. Liver Int. 2009; 29:74–81.
13. D'Souza R, Foster GR. Diagnosis and treatment of hepatitis C. J R Soc Med. 2004; 97:223–225.
14. Reichard O, Norkrans G, Frydén A, Braconier JH, Sönnerborg A, Weiland O. Randomised, double-blind, placebo-controlled trial of interferon alpha-2b with and without ribavirin for chronic hepatitis C: the Swedish Study Group. Lancet. 1998; 351:83–87.
15. Brok J, Gluud LL, Gluud C. Ribavirin monotherapy for chronic hepatitis C infection: a Cochrane Hepato-Biliary Group systematic review and meta-analysis of randomized trials. Am J Gastroenterol. 2006; 101:842–847.
16. Krishnan SM, Dixit NM. Ribavirin-induced anemia in hepatitis C virus patients undergoing combination therapy. PLoS Comput Biol. 2011; 7:e1001072.
17. Russmann S, Grattagliano I, Portincasa P, Palmieri VO, Palasciano G. Ribavirin-induced anemia: mechanisms, risk factors and related targets for future research. Curr Med Chem. 2006; 13:3351–3357.
18. McHutchison JG, Manns M, Patel K, Poynard T, Lindsay KL, Trepo C, Dienstag J, Lee WM, Mak C, Garaud JJ, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002; 123:1061–1069.
19. Reddy KR, Nelson DR, Zeuzem S. Ribavirin: current role in the optimal clinical management of chronic hepatitis C. J Hepatol. 2009; 50:402–411.
20. De Franceschi L, Fattovich G, Turrini F, Ayi K, Brugnara C, Manzato F, Noventa F, Stanzial AM, Solero P, Corrocher R. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000; 31:997–1004.
21. Takaki S, Tsubota A, Hosaka T, Akuta N, Someya T, Kobayashi M, Suzuki F, Suzuki Y, Saitoh S, Arase Y, et al. Factors contributing to ribavirin dose reduction due to anemia during interferon alfa2b and ribavirin combination therapy for chronic hepatitis C. J Gastroenterol. 2004; 39:668–673.
22. Bierau J, Lindhout M, Bakker JA. Pharmacogenetic significance of inosine triphosphatase. Pharmacogenomics. 2007; 8:1221–1228.
23. Suzuki F, Suzuki Y, Akuta N, Sezaki H, Hirakawa M, Kawamura Y, Hosaka T, Kobayashi M, Saito S, Arase Y, et al. Influence of ITPA polymorphisms on decreases of hemoglobin during treatment with pegylated interferon, ribavirin, and telaprevir. Hepatology. 2011; 53:415–421.
24. Ochi H, Maekawa T, Abe H, Hayashida Y, Nakano R, Kubo M, Tsunoda T, Hayes CN, Kumada H, Nakamura Y, et al. ITPA polymorphism affects ribavirin-induced anemia and outcomes of therapy: a genome-wide study of Japanese HCV virus patients. Gastroenterology. 2010; 139:1190–1197.
25. Shiffman ML, Ghany MG, Morgan TR, Wright EC, Everson GT, Lindsay KL, Lok AS, Bonkovsky HL, Di Bisceglie AM, Lee WM, et al. Impact of reducing peginterferon alfa-2a and ribavirin dose during retreatment in patients with chronic hepatitis C. Gastroenterology. 2007; 132:103–112.
26. Reddy KR, Shiffman ML, Morgan TR, Zeuzem S, Hadziyannis S, Hamzeh FM, Wright TL, Fried M. Impact of ribavirin dose reductions in hepatitis C virus genotype 1 patients completing peginterferon alfa-2a/ribavirin treatment. Clin Gastroenterol Hepatol. 2007; 5:124–129.
27. Shin HR, Hwang SY, Nam CM. The prevalence of hepatitis C virus infection in Korea: pooled analysis. J Korean Med Sci. 2005; 20:985–988.




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download