Abstract
We studied the expression of BRCA1, ERCC1, and RRM1 which play an important role in DNA repair systems in breast cancer. Immunohistochemical staining for EGFR, BRCA1, ERCC1, and RRM1 were performed by using a tissue microarray made from 230 breast cancer patients. Patients were classified into luminal A, luminal B, HER-2, and triple negative breast cancer (TNBC) types according to ER, PR, and HER-2 expression. The expression of ERCC1, RRM1, and BRCA1 were correlated (P < 0.05). The expression level of ERCC1 was the lowest in TNBC type (P = 0.031), ERCC1 negativity was more prominent in TNBC and luminal B groups than luminal A and HER-2 groups (P = 0.013). Cases with EGFR overexpression showed high expression of RRM1 and BRCA1 (P = 0.046, and 0.004, respectively). In conclusion, the expression of ERCC1 is particularly lower in TNBCs than other types of breast cancers.
Breast cancer is one of the most heterogeneous tumors presenting various histology, behavior, and outcome. There are many different treatment modalities for breast cancer, including drug therapy according to the characteristics of the tumors after surgery. Prototypes of drug therapy are tamoxifen for breast cancers that express hormone receptors and trastuzumab for those with HER-2 overexpression. However, there is no specific targeted therapy for triple negative breast cancer (TNBC) without the expression of hormone receptors or HER-2 until now.
Possible targeted therapies for TNBC include cytotoxic chemotherapy that causes interstrand breaks (platinum-based drugs) (1), PARP1 inhibitor (2), EGFR tyrosine kinase inhibitor (3), and c-kit tyrosine kinase inhibitor (3). Lately, clinical trials for cytotoxic chemotherapy that causes interstrand breaks have been conducted. Platinum-based chemotherapeutic agents cause DNA monoadducts and intrastrand cross-link (4), which induces normal cells to correct DNA adducts through DNA repair systems. However, TNBCs have abundant DNA aberration, suggesting a defective DNA repair system (5). BRCA1 is one of the important factors in this DNA repair system. It is involved in homologous recombination repair, non-homologous repair, and nucleotide excision repair, which are critical DNA repair systems (6). Hereditary BRCA1-associated breast cancer with germ line mutation of BRCA1 gene accounts for only 5% of all breast cancers (7), and BRCA1 mutation is rare in sporadic breast cancers (8).
However, there are considerable similarities in morphologic phenotypes and molecular features (e.g. ER negativity, high nuclear grade, high Ki-67 staining, CK5/6 expression, and EGFR expression) between sporadic TNBCs and BRCA1 related breast cancers related to BRCA1 mutation carrier (9, 10). Also, the possibility of dysfunctional BRCA1 pathway without BRCA1 somatic mutation in the sporadic breast cancer is raised by the findings of significantly low BRCA1 protein expression in ER(-), PR(-), and basal-like phenotypes, considerably low BRCA1 mRNA expression in basal-like sporadic breast cancers, and high level of ID4, a negative regulator of BRCA1, in sporadic basal-like breast cancers (11).
We examined the expression of BRCA1, ERCC1, and RRM1 which play an important role in DNA repair systems in breast cancer according to immunohistochemical phenotypes and investigated a possibility of their predictive role in chemotherapy that causes DNA interstrand breaks in breast cancer.
From the Department of Pathology of Severance Hospital, we retrieved tissue samples of patients with invasive ductal carcinoma of the breast, which were filed between January 2000 and December 2001. Formalin-fixed and paraffin-embedded tissue specimens from 230 patients with primary breast cancer were included. All archival hematoxylin and eosin (H&E)-stained slides for each patient were reviewed by two pathologists. The histological grade was assessed using the Nottingham grading system (12), and nuclear grade was evaluated according to the modified Black's nuclear grade (1 = low grade, 2 = intermediate grade, and 3 = high grade) (13). Histologic parameters were evaluated using the H&E-stained slides. Histologic parameters such as histologic subtype, nuclear grade and histologic grade were evaluated using H&E-stained slides. Clinical parameters included patient age at initial diagnosis, tumor stage of American Joint Committee on Cancer (AJCC) criteria, lymph node status, local recurrence, systemic recurrence, and patient survival.
On each H&E-stained slide, a representative area was selected and a corresponding spot was marked on the surface of the paraffin block. Using a biopsy needle, the selected area was punched out and a 3-mm tissue core was placed into a 6 × 5 recipient block. Two tissue cores of invasive component were extracted to minimize extraction bias. Each tissue core was assigned with a unique tissue microarray location number that was linked to a database containing other clinicopathologic data.
All immunostainings were performed using formalin-fixed, paraffin-embedded tissue sections. Five micrometer-thick sections were obtained by using a microtome, transferred to adhesive slides, and dried at 62℃ for 30 min. The sections were then placed in a glass jar with 10 mM citrate buffer (pH 6.0), and they were irradiated in a microwave oven for 15 min. The sections were allowed to cool in the jar at room temperature for 20 min. The slides were then rinsed with Tris buffered saline (TBS). A blocking reagent was added for 10 min after quenching the endogenous peroxidase activity in 0.3% hydrogen peroxide for 10 min. After incubation with primary antibodies (EGFR [EGFR.25, 1:50, Novocastra, UK], BRAC1 [MS110, 1:100, Oncogene, UK], ERCC1 [8F1, 1:100, Abcam, UK], and RRM1 [polyclonal, 1:100, Abcam, UK]), immunodetection was performed with biotinylated anti-mouse immunoglobulin, followed by peroxidase-labeled streptavidin using a labeled streptavidin biotin kit with 3,3'-diaminobenzidine chromogen as substrate. Optimal primary antibody incubation time and concentration were determined via serial dilution for each immunohistochemical assay with an identically fixed and embedded tissue block. Slides were counterstained using Harris hematoxylin. The staining was interpreted by two pathologists under a multiview microscope. Different results were discussed, and, in the case of persistent discordance, a third pathologist was consulted. EGFR staining was scored as follows: 0 for no membrane staining; 1+ for faint, partial membrane staining; 2+ for weak, complete membrane staining in > 10% of tumor cells; and 3+ for intense complete membrane staining in > 10% of tumor cells. Tumors with a score of 2+ or 3+ were interpreted as positive for overexpression.
For evaluating the expression of BRCA1, ERCC1, and RRM1, the percentage of expressed tumor cells were examined. Nuclear or cytoplasmic staining of more than 10% of the cells was considered positive expression. The percentage of expressed tumor cells was used as "expression index" in this study.
In this study, we classified breast cancer phenotypes according to the immunohistochemistry and FISH results for ER, PR, and HER-2 as follows: luminal A type, ER or/and PR positive and HER-2 negative, luminal B type, ER or/and PR positive and HER-2 overexpressed or/and amplified, HER-2 overexpression type, ER and PR negative and HER-2 overexpressed or/and amplified, and TNBC type, ER, PR, and HER-2 negative.
Data were processed using SPSS for Windows, version 12.0 (SPSS Inc., Chicago, IL, USA). Student's t and Fisher's exact tests were used to examine any difference in continuous and categorical variables, respectively. Significance was assumed when P < 0.05. Kaplan-Meier survival curves and log-rank statistics were employed to evaluate time to tumor metastasis and time to survival. Multivariate regression analysis was performed using Cox proportional hazards model.
Clinicopathologic characteristics of breast cancers in this study are summarized in Table 1. Sixty patients (26.0%) were classified as TNBCs, 28 patients (12.2%) as HER-2 types, 119 patients (51.7%) as luminal type A, and 23 patients as luminal type B (10.0%) in a total of 230 patients. Medullary carcinomas and metaplastic carcinomas were highly relevant to TNBCs (P < 0.001). TNBCs and HER-2 types show higher nuclear grade (P < 0.001), and TNBCs show higher histologic grade than other types (P < 0.001).
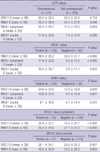
The immunohistochemical stain results of EGFR, BRCA1, ERCC1, and RRM1 are shown in Fig. 1, and Table 2. EGFR overexpression was observed in 21 patients (9.1%). Fourteen of them (66.7%) were TNBC type, and no EGFR overexpression was observed in luminal type B (P < 0.001).
ERCC1 positivity was higher in luminal type A (69.7%). In contrast, TNBC and luminal type B cancers tended to be negative for ERCC1 (51.7% and 56.5%, respectively, P = 0.013). RRM1 expression was not significantly different between the subtypes of breast cancers (P = 0.301). Cytoplasmic BRCA1 expression was noted in 84 (36.5%) cases and nuclear BRCA1 expression was noted in 57 (24.7%) cases. Cytoplasmic and nuclear BRCA1 expression were not significantly different between the subtypes of breast cancers (P = 0.320, and 0.064, respectively).
In analysis of ERCC1, RRM1, and BRCA1 by continuous variable of expression index (%), ERCC1 showed the lowest mean for TNBCs and the highest mean for luminal type A (P = 0.031), and RRM1 showed the lowest mean for luminal type A and the highest mean for HER-2 type (P = 0.001).
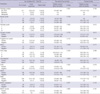
The relation among EGFR, ERCC1, RRM1, and BRCA1 expressions is shown in Table 3. RRM1 and cytoplasmic BRCA1 expression indices were higher when EGFR was overexpressed (P = 0.046 and 0.004, respectively). The positivity of ERCC1, RRM1, and cytoplasmic BRCA1 were significantly related to each other (P < 0.05, Table 3). Nuclear BRCA1 expression index was significantly higher when ERCC1 was positive (P = 0.003). Moreover, ERCC1, RRM1, and BRCA1 expression indices were significantly related (Fig. 2).
Disease free survival and overall survival according to clinicopathologic and immunohistochemical parameters are shown in Table 4. A group with lymph node metastasis showed shorter disease free survival (P = 0.009), and a group with positive estrogen receptor showed longer overall survival than negative estrogen receptor (P = 0.014). However, there was no relation among the expression of BRCA1, ERCC1, and RRM1, disease free survival, and overall survival.
In this study, EGFR overexpression was observed in 9.1% out of 230 breast cancers and in 66.7% of TNBCs (P < 0.001). This could be expected from the fact that EGFR, like cytokeratin 5/6, is known as a surrogate marker of basal-like breast cancer (3), and basal-like breast cancers are fairly overlapped with TNBCs. EGFR overexpression rate in TNBCs has been reported to be 56%-84% (14, 15), which is higher than this study. However, it is difficult to directly compare others' results with this study because different EGFR staining antibodies were used, thus different positive criteria. In this study, there was no EGFR overexpression in luminal type A and type B, but 14.3% of HER-2 type breast cancers showed EGFR overexpression in this study. About 15%-45% of basal-like breast cancers classified by microarray-based expression profiling analysis can express one of ER, PR, and HER-2 in immunohistochemical staining (3, 16, 17). Therefore, not completely coincident with TNBCs, basal-like breast cancers, which may show EGFR overexpression, can partly exist in HER-2 type.
ERCC1 negativity was higher in TNBC and luminal type B than HER-2 and luminal type A (P = 0.013), and the expression index of ERCC1 was the lowest in TNBCs (P = 0.031). In particular, TNBCs without adequate targeted therapy besides surgery until now showed higher negativity rate for ERCC1 than that of all breast cancers.
ERCC1 is an endonuclease which plays an important role in repair of DNA double strand break (18). Recently, the correlation between ERCC1 expression and repair of cisplatin-induced DNA adducts has been suggested (19, 20). ERCC1 has an ability to repair DNA adducts in the presence of platinum-based drugs such as cisplatin (21). Therefore, better therapeutic response can be expected for platinum-based drugs in tumors with low ERCC1 expression. Actually, some reports have shown better response with platinum-based chemotherapy in the case of low ERCC1 expression in human ovarian cancer cell line (22), gastric cancer (19), colorectal cancer (20), and especially, non-small cell lung cancer (23). However, to our knowledge, expression of ERCC1 in breast cancers has not been sufficiently studied (24, 25).
In this study, ERCC1 negativity rate was higher in TNBC compared to all study population, and the expression index of ERCC1 is lowest in TNBC. So, the possibility of the use of platinum-based drug as treatment modality for TNBC can be expected which was supported by a phase II clinical trial for neoadjuvant cisplatin chemotherapy in 22 TNBC patients, showing almost 64% response rate and 22% pathologically complete response (26). So, more clinical trials are needed to examine the correlation between ERCC1 expression and response to platinum-based drug therapy.
RRM1 and BRCA1 also have roles in DNA repair systems like ERCC1 does. In this study, RRM1 showed higher expression index than ERCC1 and BRCA1 in breast cancers. Previous study reported higher RRM1 protein expression than ERCC1 and BRCA1 in metastatic breast cancers (24). However, the direct comparison of the result of present study to that of previous study is difficult because previous study showed the expression range but this study demonstrated expression index. BRCA1 was negative in 58.7% of breast cancers in this study. Considering that only 5% of all breast cancers were related to BRCA1 mutation, the percentage of BRCA1 negative breast cancers in this study is very high. Previous reports on the correlation between BRCA1 gene status and BRCA1 immunohistochemistry showed that the sensitivity and specificity for BRCA1 gene mutation widely varied according to antibody type (27, 28). Since this means BRCA1 immunohistochemistry is a restrictive method to detect of BRCA1 gene mutation, it is inadequate to correlate immunohistochemical staining result for BRCA1 with BRCA1 gene mutation status.
We found that breast cancers with EGFR overexpression show higher expression rate of RRM1 and BRCA1 (P = 0.046, and 0.011, respectively), which is supported by a finding that EGFR-mediated signal pathway up-regulates DNA repair proteins (29). Also, an analysis of 84 cases with cytoplasmic expression out of 96 BRCA1 positive cases corresponds with a previous article showing a significant relation between cytoplasmic BRCA1 expression on immunohistochemistry and EGFR expression (30). Not knowing the exact mechanisms, there seems to be a correlation between altered BRCA1 expression and EGFR overexpression. In addition, the altered BRCA1 expression was associated with shorter disease free survival (30). We found a significant correlation between ERCC1, RRM1 and BRCA1 expression in this study, which is consistent with previous reports (24, 25). Defective BRCA1 genes caused up-regulation of RAD52 and XRCC4, and down-regulation of ERCC1 and RRM1 as a kind of compensation (25).
In conclusion, the expression of ERCC1, RRM1, and BRCA1 are closely associated with one another in breast cancers. The expression of ERCC1 is particularly lower in TNBCs than other types of breast cancers.
Figures and Tables
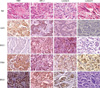
Fig. 1
EGFR, ERCC1, RRM1, and BRCA1 immunohistochemistry result according to breast cancer phenotype. EGFR overexpression is noted in TNBC and HER-2 type. ERCC1 negativity is observed in TNBC and luminal B.
AUTHOR SUMMARY
The Expression of ERCC1, RRM1, and BRCA1 in Breast Cancer According to the Immunohistochemical Phenotypes
Dokyung Kim, Woohee Jung, and Ja Seung Koo
Hereditary breast cancer is associated with mutation of BRCA1, a DNA repair gene. We investigated the expression of DNA repair gene such as ERCC1, RRM1, and BRCA1 in breast cancer, particularly according to the immunohistochemical phenotype determined by the results of estrogen receptor, progesterone receptor, and HER-2. The triple negative breast cancer showed lower expression of ERCC1 than any other type breast cancer, which could be related to the role of ERCC1 as a predictable factor of platinum-based chemotherapy.
References
1. Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000. 275:23899–23903.
2. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005. 434:917–921.
3. Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004. 10:5367–5374.
4. Zdraveski ZZ, Mello JA, Marinus MG, Essigmann JM. Multiple pathways of recombination define cellular responses to cisplatin. Chem Biol. 2000. 7:39–50.
5. Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Børresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006. 45:1033–1040.
6. Kennedy RD, Quinn JE, Mullan PB, Johnston PG, Harkin DP. The role of BRCA1 in the cellular response to chemotherapy. J Natl Cancer Inst. 2004. 96:1659–1668.
7. Couch FJ, DeShano ML, Blackwood MA, Calzone K, Stopfer J, Campeau L, Ganguly A, Rebbeck T, Weber BL. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997. 336:1409–1415.
8. Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, Eddington K, McClure M, Frye C, Weaver-Feldhaus J, Ding W, Gholami Z, Soderkvist P, Terry L, Jhanwar S, Berchuck A, Iglehart JD, Marks J, Ballinger DG, Barrett JC, Skolnick MH, Kamb A, Wiseman R. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994. 266:120–122.
9. Foulkes WD, Brunet JS, Stefansson IM, Straume O, Chappuis PO, Bégin LR, Hamel N, Goffin JR, Wong N, Trudel M, Kapusta L, Porter P, Akslen LA. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res. 2004. 64:830–835.
10. Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004. 4:814–819.
11. Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer. 2005. 116:340–350.
12. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991. 19:403–410.
13. Cutler SJ, Black MM, Mork T, Harvei S, Freeman C. Further observations on prognostic factors in cancer of the female breast. Cancer. 1969. 24:653–667.
14. Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007. 109:25–32.
15. Tischkowitz M, Brunet JS, Bégin LR, Huntsman DG, Cheang MC, Akslen LA, Nielsen TO, Foulkes WD. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007. 7:134.
16. Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, Morandi P, Fan C, Rabiul I, Ross JS, Hortobagyi GN, Pusztai L. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005. 11:5678–5685.
17. Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003. 100:10393–10398.
18. Reardon JT, Vaisman A, Chaney SG, Sancar A. Efficient nucleotide excision repair of cisplatin, oxaliplatin, and Bis-aceto-ammine-dichloro-cyclohexylamine-platinum(IV) (JM216) platinum intrastrand DNA diadducts. Cancer Res. 1999. 59:3968–3971.
19. Metzger R, Leichman CG, Danenberg KD, Danenberg PV, Lenz HJ, Hayashi K, Groshen S, Salonga D, Cohen H, Laine L, Crookes P, Silberman H, Baranda J, Konda B, Leichman L. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol. 1998. 16:309–316.
20. Shirota Y, Stoehlmacher J, Brabender J, Xiong YP, Uetake H, Danenberg KD, Groshen S, Tsao-Wei DD, Danenberg PV, Lenz HJ. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol. 2001. 19:4298–4304.
21. Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004. 24:5776–5787.
22. Li Q, Yu JJ, Mu C, Yunmbam MK, Slavsky D, Cross CL, Bostick-Bruton F, Reed E. Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer Res. 2000. 20:645–652.
23. Ceppi P, Volante M, Novello S, Rapa I, Danenberg KD, Danenberg PV, Cambieri A, Selvaggi G, Saviozzi S, Calogero R, Papotti M, Scagliotti GV. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006. 17:1818–1825.
24. Metro G, Zheng Z, Fabi A, Schell M, Antoniani B, Mottolese M, Monteiro AN, Vici P, Lara Rivera S, Boulware D, Cognetti F, Bepler G. In situ protein expression of RRM1, ERCC1, and BRCA1 in metastatic breast cancer patients treated with gemcitabine-based chemotherapy. Cancer Invest. 2010. 28:172–180.
25. Tassone P, Di Martino MT, Ventura M, Pietragalla A, Cucinotto I, Calimeri T, Bulotta A, Neri P, Caraglia M, Tagliaferri P. Loss of BRCA1 function increases the antitumor activity of cisplatin against human breast cancer xenografts in vivo. Cancer Biol Ther. 2009. 8:648–653.
26. Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De Nicolo A, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010. 28:1145–1153.
27. Al-Mulla F, Abdulrahman M, Varadharaj G, Akhter N, Anim JT. BRCA1 gene expression in breast cancer: a correlative study between real-time RT-PCR and immunohistochemistry. J Histochem Cytochem. 2005. 53:621–629.
28. Pérez-Vallés A, Martorell-Cebollada M, Nogueira-Vázquez E, García-García JA, Fuster-Diana E. The usefulness of antibodies to the BRCA1 protein in detecting the mutated BRCA1 gene. An immunohistochemical study. J Clin Pathol. 2001. 54:476–480.
29. Yacoub A, McKinstry R, Hinman D, Chung T, Dent P, Hagan MP. Epidermal growth factor and ionizing radiation up-regulate the DNA repair genes XRCC1 and ERCC1 in DU145 and LNCaP prostate carcinoma through MAPK signaling. Radiat Res. 2003. 159:439–452.
30. Rakha EA, El-Sheikh SE, Kandil MA, El-Sayed ME, Green AR, Ellis IO. Expression of BRCA1 protein in breast cancer and its prognostic significance. Hum Pathol. 2008. 39:857–865.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download