Abstract
Although TP53 mutations have been widely studied in lung cancer, the majority of studies have focused on exons 5-8 of the gene. In addition, TP53 mutations in Korean patients with lung cancers have not been investigated. We searched for mutations in the entire coding exons, including splice sites of the gene, in Korean patients with non-small cell lung cancer (NSCLC). Mutations of the gene were determined by direct sequencing in 176 NSCLCs. Sixty-nine mutations (62 different mutations) were identified in 65 tumors. Of the 62 mutations, 12 were novel mutations. TP53 mutations were more frequent in males, ever-smokers and squamous cell carcinomas than in females, never-smokers and adenocarcinomas, respectively (all comparisons, P<0.001). Missense mutations were most common (52.2%), but frameshift, nonsense, and splice-site mutations were frequently observed at frequencies of 18.8%, 15.9% and 10.1%, respectively. Of the 69 mutations, 9 (13.0%) were found in the oligomerization domain. In addition, the proportion of mutations in the oligomerization domain was significantly higher in adenocarcinomas than in squamous cell carcinomas (23.5% vs. 2.9%, P=0.01). Our study provides clinical and molecular characteristics of TP53 mutations in Korean patients with NSCLCs.
The tumor suppressor gene, TP53, is located on chromosome 17p13 and regulates cell cycle checkpoints, DNA repair, and apoptosis, and plays a key role in carcinogenesis (1). The TP53 gene comprises 11 exons, of which exons 2-11 encode a transcription factor of 393 amino acids. The TP53 protein contains distinct functional domains: the N-terminus transactivation domain (TAD, amino acids 1-42), the sequencespecific DNA binding domain (DBD, amino acids 102-292), the oligomerization domain (OD, amino acids 323-356), and the C-terminus negative regulatory domain (amino acids 363-393). Somatic mutations in the TP53 gene are frequent in most human cancers, ranging between 5% and 80%, depending on the type, stage, and etiology of the tumors (2-4).
TP53 mutational status has been widely studied in lung cancers. It has been reported that TP53 mutations occur in approximately 70% of small cell lung cancers and ≥50% of non-small cell lung cancers (NSCLCs) (2-5). However, the frequency of TP53 mutations in lung cancers varies considerably among different populations, ranging 20-60% (6-9). Moreover, the frequency and profile of TP53 mutations are different according to the gene region examined. The pioneer studies on TP53 mutations, in which TP53 mutations are localized predominantly in exons 5-8 (10, 11), led to a bias in TP53 mutation analysis, as ≥80% of TP53 mutation studies focused on exons 5-8 (amino acids 126-306) (12, 13). However, a meta-analysis of the studies that screened the entire TP53 gene showed that a total of 13.6% of mutations occurred outside exons 5-8, indicating that the entire coding region of TP53 should be analyzed (12).
Epidemiologic characteristics of lung cancer in Korea are remarkably different form Western countries. In Korea, lung cancer occurs predominantly in male smokers, and the ratio of adenocarcinoma (AC) to squamous cell carcinoma (SCC) is relatively low (1.12), which may be due to a very high smoking rate among males (87.3%) and a low smoking rate among females (20.3%) (14). In addition, several studies have shown that the prevalence of mutations in certain genes, such as epidermal growth factor receptor (EGFR), KRAS, and LKB1 genes, varies depending on the patient's ethnicity (15, 16). Considering the differences in genetic and environmental factors related to lung cancer, it is possible that the TP53 mutation profile in Korean patients with lung cancer may be different from patients in Western countries. Therefore, we searched for TP53 mutations in Korean patients with NSCLCs and correlated the results with clinicopathologic features. This is the first study of TP53 mutations that examined all coding exons and splice sites in Korean patients with lung cancers.
Primary lung tumors and matching non-malignant lung tissues were obtained from 176 NSCLC patients who underwent curative resection at Kyungpook National University Hospital (Daegu, Korea) from January 2003 to July 2007, after obtaining the approval from the Institutional Review Board of the Hospital (Approval No., 74005-263) and the patients' written informed consent. All the patients included in this study were ethnic Koreans. Patients who underwent chemotherapy or radiotherapy prior to surgery were excluded to avoid the effects on DNA. The clinicopathologic characteristics of the patients are summarized in Table 1. Histologic type of the tumors was determined according to the World Heath Organization criteria (17): 59 tumors were SCCs (33.5%) and 117 were ACs (66.5%). There were 115 males (65.3%) and 61 females (34.7%), with age at diagnosis ranging from 40 to 82 yr in the males (median age, 65) and from 37 to 79 yr in the females (median age, 62). Patients consisted of 57 never-smokers (32.4%) and 119 smokers (67.6%). Of the 117 AC cases, 54 were never-smokers (46.2%). Pathologic staging of lung cancers was determined according to the International System for Staging Lung Cancer (18): 112 (63.6%) had stage I disease, 29 (16.5%) stage II, 35 (19.9%) stage III or IV. All of the tumor and macroscopically normal lung tissue samples were obtained at the time of surgery, and these were rapidly frozen in liquid nitrogen and stored at -80℃. Only tumors with greater than 80% of the tumor component were sent forward for DNA extraction and mutational analysis.
Genomic DNA was obtained from primary tumors and matching normal lung tissues by overnight digestion with sodium dodecyl sulfate and proteinase K (Life Technologies, Rockville, MD, USA) followed by standard phenol-chloroform (1:1) extraction and ethanol precipitation. TP53 mutational analysis of the entire coding regions, including exonintron boundaries, was performed by polymerase chain reaction (PCR) and direct sequencing. Intron-based PCR primers were designated based on the GenBank reference sequence (accession no. NT_010718). The PCR reactions were performed in a total volume of 20 µL containing 100 ng of genomic DNA, 0.2 µM of each primer, 0.2 mM dNTPs, 1 unit of Taq polymerase (Takara, Shuzo Co., Otus, Shiga, Japan), and 1×reaction buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, and 1.5 mM MgCl2). The PCR cycle conditions consisted of an initial denaturation step at 95℃ for 5 min, followed by 35 cycles at 95℃ for 30 sec; 30 sec at 53℃ to 56℃; 30 sec at 72℃; and a final elongation at 72℃ for 10 min. The primers and conditions for PCR reactions are shown in Table 2. The PCR products were purified using a GENECLEAN Turbo kit (Q-Biogene, Carlsbad, CA, USA). Sequencing was done using an ABI Prism 3100 Genetic Analyzer (PE Biosystems, Foster City, CA, USA). All sequence variants were confirmed by sequencing the products of independent PCR amplifications in both directions.
Of the 176 NSCLCs, a total of 69 mutations (62 different mutations) were detected in 65 tumors (36.9%). All the mutations found in the primary tumors were absent in DNA from the matched non-malignant lung tissues and blood lymphocytes, indicating that these alterations were somatic events. There were 4 ACs with 2 mutations: 2 had an identical silent (synonymous) mutation (c.960G>A, p.K320K in exon 9) and one missense mutation (c.1000G>T, p.G334W in exon 10; or c.854A>T, p.E285V in exon 8, for each); 1 had a missense and a nonsense mutation (c.469G>T, p.V157F in exon 5; and c.1024C>T, p.R342X in exon 10); and 1 had two missense mutations (c.374C>T, p.T125M in exon 4; and c.1009C>T, p.R337C in exon 10). When compared to the International Agency for Research on Cancer (IARC) mutation database (http://www-p53.iarc.fr), 25 of the 62 mutations were novel mutations for lung cancer: 12 frameshift mutations, 8 missense mutations, 1 nonsense mutation, 3 splice-site mutations, and 1 silent mutation. In addition, 12 of the 25 mutations were novel mutations which have not been reported in any types of cancers. Table 3 details mutations for each mutation-positive case. In addition to the somatic mutations, we observed 8 germ-line variants that likely represent polymorphisms because they were found in matched non-malignant lung tissues and blood lymphocytes (rs1800369C>T [D21D], minor allele frequency [MAF]=0.392; rs1042522C>G vR-72P], MAF=0.392; rs12947788A>G [IVS7+72], MAF=0.301; rs12931053A>C [IVS7+92], MAF=0.301; rs1788-2252G>A [E339K], MAF=0.006; IVS2+14T>C, MAF=0.006; c.91G>A [V31I], MAF=0.020; and IVS3=27insGG, MAF=0.014).
TP53 mutations were significantly more frequent in males than in females (49.6% vs. 13.1%), in ever-smokers than in never-smokers (49.6% vs. 10.5%), and in SCCs than in ACs (59.3% vs. 25.6%: all comparisons, P<0.001; Table 4). However, TP53 mutations were not associated with age and pathologic stage of the tumors.
Missense mutations were the most common type (36/69 [52.2%]). However, frameshift, nonsense, and splice-site mutations were also commonly detected at frequencies of 18.8% (13/69), 15.9% (11/69), and 10.1% (7/69), respectively. There was no significant difference in mutation type according to tumor histology. When compared to the IARC mutation database, missense mutations were less frequent, whereas non-sense and frameshift mutations were more frequent (Table 5). Missense mutations occurred predominantly (31/36 [86.1%]) in the exons coding for the DBD (a part of exon 4, and exons 5-8), whereas frameshift or nonsense mutations were scattered over the entire coding exons of the gene (Fig. 1).
The TP53 mutation pattern is shown in Fig. 2. G:C>T:A transversions were the most common type (16/69 [23.2%]), followed by del or ins (13/69 [18.8%]), G:C>A:T transitions at non-CpG sites (11/69 [15.9%]), A:T>G:C transitions (9/69 [13.0%]), and G:C>A:T transitions at CpG sites (8/69 [11.6%]). G:C>T:A transversions were more frequent in eversmokers than in never-smokers (24.2% vs. 14.3%), whereas G:C>A:T transitions at CpG sites were more frequent in never-smokers than in ever-smokers (28.6% vs. 9.7%): however, the differences were not statistically significant. In addition, no significant difference in the mutation pattern according to tumor histology was found.
Of the 69 mutations observed, 3 (4.3%) occurred in the exons coding for the TAD, 46 (66.7%; 2 in exon 4, and 44 in exons 5-8) occurred in the exons coding for the DBD, 9 (13.0%) occurred in the exon 10 coding for the OD, and 11 (15.9%) occurred in splice sites or exons outside 3 major functional domains (Table 6). The proportion of mutations in the exons coding for the DBD was not significantly different according to tumor histology (74.3% in SCCs vs. 58.8% in ACs, P=0.17). However, the proportion of mutations in the exon coding for the OD was significantly higher in ACs than in SCCs (23.5% vs 2.9%, P=0.01).
The codons with the highest mutation rates were 193, 285, and 334 (3/69 [4.3% for each]), and followed by 125, 154, 158, 214, 273, 320, 342, and 349 (2/69 [2.9% for each]). Two of these, that is, codons 158 and 273, are major mutation hot spots in lung cancer (Fig. 1). Mutations in codons 334, 342, and 349 were observed only in ACs.
We described here for the first time the TP53 mutation profile of Korean patients with NSCLCs. In the present study, we searched for TP53 mutations in entire coding exons including splice sites. In agreement with previous studies (4-7) and the IARC mutation database, TP53 mutations were significantly related to gender, smoking history, and tumor histology. However, we report notable findings in TP53 mutation analysis. First, a total of 36.2% (25/69) of the TP53 mutations occurred outside exons 5-8. Second, although missense mutations were the predominant type, frameshift, nonsense, and splice-site mutations were also frequent and they comprised 44.9% (31/69) of the TP53 mutations observed. Third, the proportion of mutations in the exon coding for the OD were significantly higher in the ACs than in the SCCs. Fourth, in the present study, TP53 mutations were detected in 59.3% of SCCs and in 25.6% of ACs. These frequencies were compatible with those of the previous studies (4-7) and the IARC mutation database. Finally, we report 12 novel mutations which have not been reported in any types of cancers.
Analysis of the studies that screened the entire coding exons of the TP53 gene showed that focusing on exons 5-8 led to an unacceptable bias (12, 13). The studies that focused on exons 5-8 have reported that most TP53 mutations were point missense mutations and predominantly occurred in the DBD. However, the studies that examined the entire TP53 gene have shown that a total of 13.6% of mutations were located outside exons 5-8, particularly in exons 4, 9, and 10 and that in the exons outside exons 5-8, nonsense and frameshift mutations were more frequent than in exons 5-8 (4, 19, 20). In the present study, 25 (36.2%) of all the 69 mutations were located outside exons 5-8. In addition, 18 (29.0%) of the 62 mutations detected in coding exons occurred outside exons 5-8 (1 in exon 3, 6 in exon 4, 2 in exon 9, and 9 in exon 10), which was significantly higher than the proportion (13.6%) in a meta-analysis of the studies that screened the entire coding exons of the TP53 gene (12).
Another possible source of bias is related to splice-site mutations. Because splice-site mutations have been thought to be relatively infrequent and their effects have not been well-characterized, most previous studies have not analyzed exonintron junctions of the gene. Therefore, in the IARC database, splice-site mutations account for 2.28% and 2.70% of all the mutations reported in SCCs and ACs of the lung, respectively (Table 5). However, Varley et al. (21) recently reported that germ-line splice-site mutations in 7 (17.5%) of 40 families with Li-Fraumeni syndrome, and splicing was altered in 6 of the 7 cases with germ-line splice-site mutations. This finding suggests the real-incidence of splice-site mutations in sporadic human cancers is closer to this figure (12). In agreement with this suggestion, 7 splice-site mutations were observed in the present study, and they comprised 10.1% of all mutations detected. In addition, 2 of the 7 splice-site mutations were novel mutations that have not been reported in any types of human cancer.
In the present study, TP53 mutations were found in 59.3% of SCCs and in 25.6% of ACs, which were compatible with prior studies (4-7) and the IARC database. However, considering that most of the prior studies have only examined exons 5-8, our findings suggest that the frequency of the TP53 mutation might be relatively low in Korean NSCLCs. The difference in the mutation frequency, as well as the difference in the mutation spectrum between patients with NSCLCs from the Korean population and other ethnic populations may be due to environmental and ethnic differences.
The TP53 protein functions as a tetramer and contains TAD that is involved in transcriptional function, DBD, and OD that permits oligomerization of the protein (22). It has been demonstrated that the OD plays an important role in DNA binding, protein-protein interactions, post-translational modifications, and TP53 degradation (23). Moreover, mutations in the OD can inactivate wild-type TP53 protein upon a dominant negative effect as mutations in the DBD that result in a dominant negative effect by altering the conformation or by decreasing the affinity for DNA (23). Several studies have reported that the OD is not often mutated in human cancer, even in the study in which the entire TP53 gene was analyzed (24, 25), therefore little attention has been given to the OD. However, in the present study, 9 (13.0%) of the 69 mutations detected occurred in the OD, suggesting that mutations in the OD occur relatively frequently in patients with lung cancer.
In conclusion, this study provides clinical and molecular characteristics of TP53 mutations in Korean patients with NSCLCs. This study shows that TP53 mutation profiles are different from those of other ethnic populations. Our results, may therefore contribute to a better understanding of lung tumorigenesis in Koreans.
Figures and Tables
Fig. 1
Overview of 69 TP53 mutations. E2-E11 do not reflect the real size of each exon, but are approximately proportional to the frequency of mutations. The position of each bar indicates the approximate location of each mutation. The height of each bar correlates with the frequency of the mutation at each location.
*1, P128_A129 del; *2, P177_C182 del; *3, I195_N200 del.
E, exon; TAD, transactivation domain; DBD, DNA-binding domain; OD, oligomerization domain.
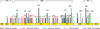
Fig. 2
TP53 mutation pattern.
SCC, squamous cell carcinoma; AC, adenocarcinoma. Number in parentheses, percentage.

References
2. Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000. 77:81–137.

3. Bennett WP, Hussain SP, Vahakangas KH, Khan MA, Shields PG, Harris CC. Molecular epidemiology of human cancer risk: gene-environemt interactions and p53 mutation spectrum in human lung cancer. J Pathol. 1999. 187:8–18.
4. Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994. 54:4855–4878.
5. Toyooka S, Tsuda T, Gazdar AF. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat. 2003. 21:229–239.
6. de Anta JM, Jassem E, Rosell R, Martinez-Roca M, Jassem J, Martinez-Lopez E, Monzo M, Sanchez-Hernandez JJ, Moreno I, Sanchez-Cespedes M. TP53 mutational pattern in Spanish and Polish non-small cell lung cancer patients: null mutations are associated with poor prognosis. Oncogene. 1997. 15:2951–2958.

7. Kishimoto Y, Murakami Y, Shiraishi M, Hayashi K, Sekiya T. Aberrations of the p53 tumor suppressor gene in human non-small cell carcinomas of the lung. Cancer Res. 1992. 52:4799–4804.
8. Ryberg D, Kure E, Lystad S, Skaug V, Stangeland L, Mercy I, Borresen AL, Haugen A. p53 mutations in lung tumors: relationship to putative susceptibility markers for cancer. Cancer Res. 1994. 54:1551–1555.

9. Takagi Y, Koo LC, Osada H, Ueda R, Kyaw K, Ma CC, Suyama M, Saji S, Takahashi T, Tominaga S, Takahashi T. Distinct mutational spectrum of the p53 gene in lung cancers from Chinese women in Hong Kong. Cancer Res. 1995. 55:5354–5357.
10. Takahashi T, Nau MM, Chiba I, Birrer MJ, Rosenberg RK, Vinocour M, Levitt M, Pass H, Gazdar AF, Minna JD. p53; a frequent target for genetic abnormalities in lung cancer. Science. 1989. 246:491–494.

11. Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P, Glover T, Collins FS, Weslon A, Modali R, Harris CC, Vogelstein B. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989. 342:705–708.

12. Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001. 1:233–240.

13. Soussi T, Lozano G. p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun. 2005. 331:834–842.

14. In KH, Kwon YS, Oh IJ, Kim KS, Jung MH, Lee KH, Kim SY, Ryu JS, Lee SY, Jeong ET, Lee SY, Yum HK, Lee CG, Kim WS, Zo JI, Kim H, Kim YW, Kim SK, Lee JC, Kim YC. Lung cancer patients who are asymptomatic at diagnosis show favorable prognosis: a Korean lung cancer registry study. Lung Cancer. 2009. 64:232–237.

15. Bae NC, Chae MH, Lee MH, Kim KM, Lee EB, Kim CH, Park TI, Han SB, Jheon S, Jung TH, Park JY. EGFR, ERBB2, and KRAS mutations in Korean non-small cell lung cancer patients. Cancer Genet Cytogenet. 2007. 173:107–113.

16. Koivunen JP, Kim J, Lee J, Rogers AM, Park JO, Zhao X, Naoki K, Okamoto I, Nakagawa K, Yeap BY, Meyerson M, Wong KK, Richards WG, Sugarbaker DJ, Johnson BE, Janne PA. Mutations in the LKB1 tumor suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008. 99:245–252.
17. Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001. 18:1059–1068.

18. Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997. 111:1710–1717.

19. Greenblatt MS, Grollman AP, Harris CC. Deletions and insertions in the p53 tumor suppressor gene in human cancers: confirmation of the DNA polymerase slippage/misalignment model. Cancer Res. 1996. 56:2130–2136.
20. Hussain SP, Harris CC. p53 mutation spectrum and load: the generation of hypotheses linking the exposure of endogenous or exogenous carcinogens to human cancer. Mutat Res. 1999. 428:23–32.

21. Varley JM, Attwooll C, White G, McGown G, Thorncroft M, Kelsey AM, Greaves M, Boyle J, Birch JM. Characterization of germline TP53 splicing mutations and their genetic and functional analysis. Oncogene. 2001. 20:2647–2654.

22. Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008. 77:557–582.

24. Soussi T, Dehouche K, Beroud C. p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum Mutat. 2000. 15:105–113.

25. Levine AJ, Wu MC, Chang A, Silver A, Attiyeh EF, Lin J, Epstein CB. The spectrum of mutations at the p53 locus. Evidence for tissue-specific mutagenesis, selection of mutant alleles, and a "gain of function" phenotype. Ann N Y Acad Sci. 1995. 768:111–128.




 PDF
PDF ePub
ePub Citation
Citation Print
Print








 XML Download
XML Download