Abstract
The pre-diagnostic test for preimplantation genetic diagnosis (PGD) of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency was performed by polymerase chain reaction (PCR) and direct sequencing for hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (HADHA) gene. We obtained unexpected genotyping results of HADHA gene by allele drop-out in the analysis of patients' genomic DNA samples with a referred PCR primer set. Upon further analysis with a re-designed primer set, we found a novel single nucleotide polymorphism (SNP) at the referred primer-binding site in the normal allele of HADHA gene (NT_022184, 5233296 a>t). We found that the frequency of this novel SNP was 0.064 in Korean population. Pre-diagnostic test using single lymphocytes and clinical PGD were successfully performed with the re-designed primer set. Nineteen embryos (95.0%) among 20 were successfully diagnosed to 5 homozygous mutated, 8 heterozygous carrier and 6 wild type. Among 6 normal embryos, well developed and selected 4 embryos were transferred into the mother's uterus, but a pregnancy was not achieved. We proposed that an unknown SNP at primer-binding sites would be a major cause of allele drop-out in the PGD for single gene disorder.
Recently, preimplantation genetic diagnosis (PGD) has been successfully applied as an alternative to prenatal diagnosis of chromosomal abnormalities and inherited diseases. The procedures for PGD involve in vitro fertilization, embryo culture, biopsy and analysis of the blastomere from preimplantation embryos using fluorescent in situ hybridization (FISH) for chromosomal alterations or polymerase chain reaction (PCR) for specific gene defects (1, 2). In case of PCR-PGD, the minuscule amount of the DNA of a single blastomere was faced with three major problems such as allele drop-out (ADO), amplification failure and contamination (3, 4). Among them, amplification failure and contamination could be solved by the optimization of PCR-PGD protocol.
The ADO is a significant and unique problem which is the failure of PCR to amplify one of the two alleles present in the DNA sample. While it occurs occasionally, no method has yet been developed to eliminate ADO in single cell analysis (5). The results by ADO may lead to misdiagnoses of heterozygous embryos as homozygous affected or unaffected embryos in PCR-PGD. Therefore, several strategies have been developed to decrease ADO, such as increasing the denaturation temperature and annealing time (6), using a more powerful lysis method (7, 8), or applying the multiplex PCR and fluorescent PCR for mutations or linked markers (9, 10).
The long-chain 3-hydroxyacyl CoA dehydrogenase (LCHAD) (EC 1.1.1.211) is a component of the tri-functional protein of the inner mitochondrial membrane with alpha and beta subunits. The LCHAD deficiency is a fatal autosomal recessive metabolic disorder and caused by mutation in the hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (HADHA) gene. Deficiency of this enzyme causes sudden unexplained infant death, a Reye-like syndrome, cardiomyopathy, or skeletal myopathy. Patients with the LCHAD deficiency may also develop long-term complications such as peripheral neuropathy or pigmentary retinopathy leading to impaired vision. The incidence of LCHAD deficiency in the general population is approximately 1 in 70,000 (11, 12). In our pre-diagnostic test for a LCHAD deficiency case, unexpected genotyping results were observed from a female patient and her daughter DNA samples with a referred primer set. The problem was settled by further genetic analysis with a re-designed primer set for the HADHA gene.
In this study, we described the process of identification of a novel single nucleotide polymorphism (SNP) at a referred primer-binding site during the pre-diagnostic test for PGD of LCHAD deficiency. We found a novel SNP of the HADHA gene and investigated the allele frequency in Korean population.
A couple (husband: 37 yr, wife: 33 yr) was referred to our center after five neonatal deaths due to severe lactic acidemia (Fig. 1). There was no significant abnormality in their medical history and physical examination. Molecular genetic analysis of the couple's HADHA gene was performed by PCR and direct sequencing in Seoul Asan Hospital, and revealed a single base substitution (IVS16+2 t>g) in the intron 16 of HADHA gene resulting in total deletion of exon 16. Unfortunately, these mis-splicing mutations were detected in all family members. They are carriers for IVS16+2 t>g in the HADHA gene. The couple requested PGD for a pregnancy with an unaffected baby.
The pre-diagnostic test for PCR-PGD for the single gene disorder was composed of two steps in our center. First step was to confirm the reported genotyping with genomic DNA from patients and normal relatives. Second step was to optimize our protocol for single blastomere analysis using single lymphocytes samples. Genomic DNA was extracted from 5 mL EDTA anti-coagulated blood samples using the AquaPure Genomic DNA Isolation kit (Bio-Rad Laboratories, Hercules, CA, U.S.A.) according to the manufacturer's instructions.
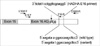
We used two kinds of primer sets for the pre-diagnostic test of LCHAD deficiency in this study. One was a referred primer set (HADHA-E16), which was applied for screening the mutation, at Seoul Asan Hospital. Another was a re-designed primer set (HADHA-E16W, Table 1, Fig. 2) by our center which amplified the outer region of the referred primer. The DNA fragments from PCR were analyzed by the direct DNA sequencing as described below.
In order to determine the efficiency of single cell PCR and ADO rates, the pre-diagnostic testing was performed with single lymphocytes and the primers, which were used for the detection of the causative mutation. Lymphocytes were isolated from 10 mL of peripheral blood in heparinized tubes collected from the female patient and her husband using Ficoll-Paque density gradient separation (Ficoll-Paque™PLUS, Amersham Biosciences AB, Uppsala, Sweden) according to the manufacturer's protocol. The cell layer containing lymphocytes was removed and diluted with sterile and filtered phosphate-buffered saline (PBS) to adjust cell density for single cell isolation. Single lymphocytes were isolated using a fine heat-polished glass micropipette and then loaded into 0.2 mL thin wall PCR tubes containing 5 µL of alkaline lysis buffer (200 mM/L KOH and 50 mM/L dithiothreitol) under a stereo microscope. The single lymphocyte samples were stored at -20℃ until analysis.
Controlled ovarian hyperstimulation was done as previously described (1). Following ovarian stimulation, follicles were aspirated and matured oocytes were fertilized by intracytoplasmic sperm injection (ICSI). Fertilized oocytes were cultured in the G1.2 medium (Vitrolife Sweden AB, Kungsbacka, Sweden) for three days. On the third day of culture, embryos were incubated for 5 min in Ca2+/Mg2+-free medium (EB-10™, Vitrolife Sweden AB) before biopsy. The acid Tyrode's solution (ZD-10™, Vitrolife Sweden AB) was applied to create a small hole in the zona pellucida. Biopsy was performed by gentle aspiration using a polished micropipette with 30 µm of inner diameter. One blastomere was biopsied on embryos with less than 6-cells and two blastomeres on six or more cells. After the blastomere biopsy procedure, embryos were washed several times and transferred to the G2.2 medium (Vitrolife Sweden AB). Each blastomere was washed twice through two drops of G2.2 medium and transferred into sterile 0.2 mL PCR tubes containing 5 µL of alkaline lysis buffer. For each biopsied blastomere, blank negative controls were prepared from the washing drops. Embryos were cultured under the standard culture condition until the diagnosis was accomplished. Embryos with normal genotype were selected and transferred into the mother's uterus on the fourth day of culture.
The lymphocytes and blastomeres were prepared with alkaline lysis buffer at 65℃ for 10 min incubation. The buffer containing sample was neutralized by the addition of 5 µL of neutralization buffer (900 mM/L Tris-HCl, 300 mM/L KCl, 200 mM/L HCl) before proceeding to PCR. The PCR strategy consisted of the first PCR followed by the hemi-nested PCR for the mutation of the HADHA gene. After cell lysis and neutralization, 1.5 mM/L MgCl2, 200 µM/L of each dNTP (Roche Diagnostics GmbH, Mannheim, Germany), 1 IU Supertherm DNA polymerase (JMR HOLDINGS, London, U.K.) and 0.5 µM/L of each outer primers, were added to each tube for a total volume of 30 µL. The first round of PCR involved a 96℃ denaturation temperature for 10 min as a means to reduce ADO, followed by 25 cycles consisting of 95℃ for 40 sec, 62℃ for 1 min and 72℃ for 1 min, and followed by a final extension step of 10 min at 72℃ on a GeneAmp PCR System 2700 (Applied Biosystems, Foster City, CA, U.S.A.). For the second round of DNA amplification, 1 µL of the first PCR reaction products was added to another tube containing 2 µL of PCR buffer (50 mM/L KCl, 10 mM/L Tris-HCl, pH 8.3), 1.5 mM/L MgCl2, 200 µM/L of each dNTP, 2 IU SynergyN DNA polymerase (GeneCraft Co, Munster, Germany) and 0.5 µM/L of each inner primer, and the amplification was performed with 40 cycles as described above. In order to detect PCR products, 3 µL of each PCR product was subjected to electrophoresis in the 2% agarose gel.
For DNA sequence analyses, the PCR products were purified by the purification kit (Qiagen GmbH, Hilden, Germany). The purified PCR products (40 ng) were sequenced by direct cycle sequencing using fluorescent-labelled dideoxy terminators (Big Dye Terminator Cycle Sequencing Ready Reaction Kit; Applied Biosystems, Foster City, CA, U.S.A.), according to the manufacturer's protocol. The reaction conditions were as follows: 25 PCR cycles, a denaturation step of 10 sec at 96℃, annealing for 5 sec at 50℃ and extension for 4 min at 60℃ on a GeneAmp PCR System 2700 (Applied Biosystems). The samples were then cleared using ethanol precipitation for unincorporated dye terminator removal. The precipitated pellets were resuspended with 20 µL of Hi-Di Formamide (Applied Biosystems), denatured at 95℃ for 4 min and run on ABI Prism 3100 Avant automated genetic analyzer (Applied Biosystems). The sequences were compared with the wild type controls using Seqscape Software (Applied Biosystems) for mutation analysis.
To determine the frequency of a novel SNP of the HADHA gene in the Korean population, genomic DNA samples from unrelated individuals (n=63) were extracted, then analyzed by PCR and direct sequencing as described above. This study was approved by the Institutional Review Board (IRB) as SCH-IRB-2004-01. Written consent was obtained from the subjects in order to confirm that genomic DNA could be used for research purposes.
Before the clinical trial of PGD, we always confirm the mutation site of parents and their children or relatives with individual's genomic DNA samples, and then perform the single lymphocyte analysis. PCR amplification was carried out with the referred primers, which were designed for genetic screening at Seoul Asan Hospital, and then the DNA fragments of PCR was carried out direct sequencing. However, compared with previous reports from Seoul Asan Hospital, we observed different genotype of the HADHA gene in the family. The male patient had a heterozygous genotype, the same result (carrier of HADHA IVS16+2 t>g) as the previous reports. However, the female patient and her daughter showed unexpected results. According to our results of PCR with the referred primer (HADHA-E16), the female patient and her daughter were not identified as heterozygous genotype, but homozygous for IVS16+2 t>g of the HADHA gene. And then, we performed several independent PCR analyses in different PCR conditions. However, we could not figure out any problem with the referred primer set.
To settle the problem, the unexpected genotyping of the HADHA gene for the female patient and her daughter, DNA samples were extracted again and primers were re-designed to amplify a larger region containing HADHA-E16 primer-binding sites (Table 1, Fig. 2). After the amplification with the re-designed primer set (HADHA-E16W) and direct sequencing of PCR products, both patients and their daughter were confirmed as heterozygotes for the mutated site.
As results of complete sequencing of both strands for PCR products amplified with HADHA-E16W primers, a novel SNP was found at the binding site of the referred reverse primer for the HADHA gene. We registered the novel SNP of the HADHA gene on the GenBank site (NT_022184, 5233296 a>t NCBI refSNP ID: rs17549879). The novel SNP is located at the 7th position of the 3' end of the referred reverse primer sequences in only normal alleles of the female patient and her daughter (Table 1, Fig. 3). However, the normal allele of the male patient had wild type sequence for this site. This a/t polymorphism near the 3' end of the referred reverse primer was the very reason that the normal allele of the female patient and her daughter was dropped out in the PCR with the referred primer.
We evaluated the frequency of the SNP in 63 unrelated individuals from the Korean population. Eight individuals (12.7%) were found to have this novel SNP as heterozygous types. There was no homozygous type for this SNP in tested samples. The SNP frequency of the HADAH gene in Korean population was determined to be 0.064 by studying 126 chromosomes. This SNP was in Hardy-Weinberg equilibrium (χ2=0.290, df=1).
The efficiency and accuracy of the single cell PCR analysis was tested using single lymphocytes of peripheral blood from the male and female patient with heterozygous genotype. PCR on single lymphocytes resulted in 95.9% (70/73) amplification rate, whereas none of the negative controls showed a band. The ADO of single lymphocytes PCR by direct DNA sequencing was detected in 16 out of 70 lymphocytes (22.9%) in the PCR and direct sequencing.
After ovarian hyperstimulation, a total of 25 oocytes were retrieved and 20 matured oocytes (metaphase-II) were iseminated by ICSI, and then fertilized 20 embryos were diagnosed. Two blastomeres were biopsied from 14 embryos and one blastomere from 6 embryos, respectively. In the PCR and direct sequencing, 33 blastomeres of 19 embryos among 34 blastomeres of 20 embryos were successfully amplified and diagnosed. Amplification rate per blastomere and embryo was 97.1% and 95.0%, respectively. There were no amplifications in all negative controls. The ADO of single cell PCR was detected in 7 of 33 amplified samples (21.2%). Nineteen embryos (95.0%) among 20 were successfully diagnosed to 5 homozygous mutated, 8 heterozygous carrier and 6 wild type. Among 6 normal embryos, well developed and selected 4 embryos were transferred into the mother's uterus, but a pregnancy was not achieved.
In this study, a novel SNP at the primer-binding site was identified by PCR with a newly re-designed primer set and direct sequencing. The frequency of this novel SNP was 0.064 in Korean population of this study. Although we have not tested populations of other nations, any frequency above 0.01 is considered polymorphic in a population. We registered this novel polymorphism at the GenBank as NT_022184, 5233296 a>t, NCBI refSNP ID: rs17549879. The novel SNP might be a major cause of amplification failure of the normal allele in the female patient and her daughter, thus only mutated allele of the HADHA gene was amplified with the referred primer in our stringent PCR condition of PGD.
In our pre-diagnostic test with genomic DNA samples, the discrepancy of genotyping of the HADHA gene using the referred primer set occurred between our center and another mutation screening laboratory. We have discussed about this problem several times hoping to find a solution. They used a different Taq. polymerase and PCR protocols from our high stringent PCR condition for PGD, which was important to prevent the generation of non-specific PCR products. Our laboratory has endeavored to optimize single cell PCR conditions for PGD including longer initial denaturation time, higher denaturation temperature, a specific Taq polymerase, adjust of MgCl2 concentration and alkaline lysis methods (8). Several independent PCR analyses were tried using genomic DNA samples by our stringent PCR condition, however, we could not figure out the problem with the referred primer set. Upon further genetic analysis with a re-designed primer set, we found a novel SNP of the HADHA gene at the binding site of the referred primer in the normal allele of the female patient and her daughter. With the re-designed primer set, pre-diagnostic tests in single lymphocytes and clinical PGD trial were successfully done.
Most researchers for PGD make an effort to optimize the PCR conditions with single cell samples. In this respect, our study shows the importance of thoroughly characterizing the performance of primer sets prior to changing amplification conditions or DNA polymerases. The distinct genotyping for a mutation in a single gene disorder, especially autosomal dominant disorder, could be inconsistent with the expected results. When this situation occurs, the possibility of SNP at the primer-binding site should be speculated. Although none of the subjects in this study carried SNP on the mutant allele, if SNP was on the mutant allele, the mutant allele could be dropped-out. The presence of the unidentified SNP may lead to incorrect genotyping and potential misdiagnosis in clinical PGD cases. Therefore, it is very important to test a significant number of individuals with used primers to evaluate the SNP in the primer binding sites. Several reports showed the potential error in molecular diagnosis from unidentified SNPs and mutations like this study (13-16).
In conclusion, we report the successful PGD in a couple at risk of transmitting the LCHAD deficiency, although no pregnancy resulted. As far as we know, this is the second report in the world and the first report in Korea that successful PGD for LCHAD deficiency case (17). We identified the novel SNP of the HADHA gene in the referred primer-binding site, and it was the major cause of ADO for the normal allele in the pre-diagnostic tests for PGD of LCHAD deficiency. We proposed that an unknown SNP at primer-binding sites would be a major cause of allele drop-out in the PGD for single gene disorder.
Figures and Tables
Fig. 1
Pedigree of the family having LCHAD deficiency. Father, mother and their live daughter have both wild (W) and mutant (M) allele of the HADHA gene. Father and mother have the same HADHA gene mutation site (♦). Both mother and her daughter have a normal allele with an unidentified SNP (▴).
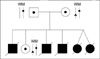
Fig. 2
Schematic representation for a part of the HADHA gene, together with the position of SNP and two pairs of primers used in this study. PCR primer sets were designed from the 5' and 3' flanking regions of the exon 16 in the HADHA gene. The right uppermost is the sequences of reverse primer (E16-R) and two lowers are the sequences of genomic DNA of wild and variant type alleles, respectively. The horizontal arrows indicate the direction and the location of each primer and the vertical arrow indicates the mutation loci (HADHA IVS16+2 t>g) in this family.
References
1. Lim CK, Jun JH, Min DM, Lee HS, Kim JY, Koong MK, Kang IS. Efficacy and clinical outcome of preimplantation genetic diagnosis using FISH for couples of reciprocal and Robertsonian translocations: the Korean experience. Prenat Diagn. 2004. 24:556–561.
2. Lee HS, Choi HW, Lim CK, Min DM, Byun HK, Kim JY, Koong MK, Yoo HW, Kim SC, Jun JH, Kang IS. Successful preimplantation genetic diagnosis for ornithine transcarbamylase deficiency, junctional epidermolysis bullosa and lactic acidosis using duplex nested PCR: delivery of healthy baby by specific preimplantation genetic diagnosis for ornithine transcarbamylase deficiency. Korean J Obstet Gynecol. 2004. 47:708–718.
3. Sermon K. Current concepts in preimplantation genetic diagnosis (PGD): a molecular biologist's view. Hum Reprod Update. 2002. 8:11–20.

4. Piyamongkol W, Bermudez MG, Harper JC, Wells D. Detailed investigation of factors influencing amplification efficiency and allele drop-out in single cell PCR: implications for preimplantation genetic diagnosis. Mol Hum Reprod. 2003. 9:411–420.

5. Findlay I, Ray P, Quirke P, Rutherford A, Lilford R. Allelic drop-out and preferential amplification in single cells and human blastomeres: implications for preimplantation diagnosis of sex and cystic fibrosis. Hum Reprod. 1995. 10:1609–1618.

6. Ray PF, Handyside AH. Increasing the denaturation temperature during the first cycles of amplification reduces allele dropout from single cells for preimplantation genetic diagnosis. Mol Hum Reprod. 1996. 2:213–218.
7. Thornhill AR, McGrath JA, Eady RA, Braude PR, Handyside AH. A comparison of different lysis buffers to assess allele dropout from single cells for preimplantation genetic diagnosis. Prenat Diagn. 2001. 21:490–497.

8. Choi HW, Lee HS, Lim CK, Koong MK, Kang IS, Jun JH. Reliability of the single cell PCR analysis for preimplantation genetic diagnosis of single gene disorders. Korean J Fertil Steril. 2005. 32:293–300.
9. Dreesen J, Jacobs L, Bras M, Herberg J, Dumoulin JC, Geraedts JP, Evers JL, Smeets HJ. Multiplex PCR of polymorphic markers flanking the CFTR gene: a general approach for preimplantation genetic diagnosis of cystic fibrosis. Mol Hum Reprod. 2003. 6:391–396.

10. Lee HS, Choi HW, Lim CK, Park SY, Kim JY, Koong MK, Jun JH, Kang IS. Efficacy of duplex-nested PCR and fluorescent PCR in the preimplantation genetic diagnosis for Duchenne muscular dystrophy. Korean J Fertil Steril. 2005. 32:17–26.
11. Strauss AW, Bennett MJ, Rinaldo P, Sims HF, O'Brien LK, Gibson B, Ibdah J. Inherited long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency and a fetal-maternal interaction cause maternal liver disease and other pregnancy complications. Semin Perinatol. 1999. 23:100–112.

12. Gillingham MB, Connor WE, Matern D, Rinaldo P, Burlingame T, Meeuws K, Harding CO. Optimal dietary theraphy of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency. Mol Genet Metab. 2003. 79:114–123.
13. Wong LJ, Chen TJ, Dai P, Bird L, Muenke M. Novel SNP at the common primer site of exon IIIa of FGFR2 gene causes error in molecular diagnosis of Craniosynostosis syndrome. Am J Med Genet. 2001. 102:282–285.
14. Solano AR, Dourisboure RJ, Weitzel J, Podesta EJ. A cautionary note: false homozyosity for BRCA2 6174delT mutation resulting from a single nucleotide polymorphism masking the wt allele. Eur J Hum Genet. 2002. 10:395–397.
15. Zajickova K, Krepelova A, Zofkova I. A single nucleotide polymorphism under the reverse primer binding site may lead to BsmI misgenotyping in the vitamin D receptor gene. J Bone Miner Res. 2003. 18:1754–1757.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download