Abstract
The hypermethylation of the CpG islands is a common mechanism for the inactivation of tumor-related genes. In the present study, we analyzed the methylation status of genes for cell repair such as hMLH1, MGMT, and GSTP1, and a gastric cancer-specifically methylated DNA fragment, MINT 25 in gastric cancer cases and control groups. The study population consisted of 100 gastric cancer patients (50 distal and 50 proximal carcinomas), and 238 healthy controls. All genes showed more frequent hypermethylation in the cases than in the control group (p<0.0001). We investigated the association between promoter hypermethylation and relevant parameters including age, gender, alcohol consumption, smoking, and family history. There was a common hypermethylation of hMLH1 (p=0.008), MGMT (p=0.0001), and GSTP1 (p=0.0003) in females. This study also demonstrates that hypermethylation was strongly associated with non-drinkers (MGMT, p=0.046 and MINT 25, p=0.049) and non-smokers (hMLH1, p=0.044; MGMT, p=0.0003; MINT 25, p=0.029). Moreover, the frequency of MINT 25 hypermethylation increased with age (p=0.037), and MGMT methylation was frequently detected in distal gastric cancer than in proximal type (p=0.038). Our study suggested that promoter hypermethylation of the genes involved in cell repair system and MINT 25 is associated strongly with some subgroups of primary gastric carcinoma.
Aberrant methylation patterns are one of the fundamental hallmarks of cancer cells. Tumor cells are generally hypomethylated relative to normal cells with regional hypermethylation. CpG islands are GC-rich areas of the genome corresponding to the promoter regions of genes and are associated with transcriptional activity (1). The methylation status of these CpG islands has been shown to be involved with oncogene activation and tumor suppressor gene inactivation. A recent study on the profile of promoter hypermethylation for 12 genes (p16INK4A, p15INK4B, p14ARF, p73, APC, BRCA1, hMLH1, GSTP1, MGMT, CDMI, TIMP3, and DAPK) in 15 major tumor types revealed that one or more of the genes are hypermethylated in all tumor types (2). The profile of the promoter hypermethylation for the genes, however, differs in each cancer type, providing a tumor type- and gene-specific profile.
Aberrant methylation in tumor-related genes is frequently detected in gastric intestinal metaplasia with and without gastric cancer, suggesting their early involvement in the multistep progression of gastric carcinogenesis (3). The identification of genes targeted by hypermethylation may provide insights into the mechanisms for the inactivation of tumor-suppressive pathways in gastric cancer cases. In addition, hypermethylated genes may serve as targets for the development of new screening tests for cancer (4).
In the present study, we compared the hypermethylation of genes responsible for the cell repair system (hMLH1, MGMT, and GSTP1) in gastric cancer case and control study. We also analyzed MINT 25 (methylated in tumors 25) which showed a high frequency of methylation in gastric carcinomas (5, 6). There are many reports that have shown frequent hypermethylation of these genes in gastric carcinoma, but its interaction with histological characteristics is still unclear. Thus, we evaluated the association between the hypermethylation of these genes and gastric cancer according to the risk factors such as H. pylori infection, alcohol consumption, smoking, family history, and their histological characteristics by the methylation-specific PCR (MSP) method.
A case-control study of gastric cancer was performed in Daegu City. One hundred gastric cancer patients and two hundred thirty-eight healthy subjects participated in this study. Patients affected with gastric cancer were considered eligible if they had histologically diagnosed adenocarcinoma of the stomach. The control group included subjects who had no current or previous diagnosis of cancer. Data included questionnaire data, and a review of medical records.
Whole blood samples of control group were used to isolate genomic DNA by the phenol-chloroform method (7). Tumor tissues were used to determine DNA methylation status of cancer patients. Frozen tumor tissues were ground and incubated at 50℃ for 3 hr in a lysis buffer containing 0.5% of sodium dodecyl sulfate (SDS), followed by a phenolchloroform method. The amount of DNA and their purity were determined by spectrophotometry.
DNA methylation patterns in the CpG islands of the target genes were determined by chemical modification of unmethylated, but not methylated, cytosines to uracils, and subsequent PCR amplification using primers specific for either methylated or modified unmethylated DNA (8). The bisulfite-modification was performed according to Olek et al. (9). One microgram of DNA was denatured by NaOH and modified by sodium bisulfite. DNA samples were then purified using Wizard DNA purification resin (Promega, Madison, WI, U.S.A.), treated with NaOH again, and precipitated with ethanol. DNA was resuspended in water and used immediately or stored at -20℃.
Two µL of treated DNA were used for each PCR reaction. MSP showed the presence or absence of methylated genes of hMLH1, MGMT, and GSTP1. The methylation status of MINT 25 was determined by bisulfite-PCR followed by restriction digestion. Two µL of DNA modified with bisulfite were amplified and 15 µL of the PCR products were then digested with RsaI, which is specific to the methylated alleles by virtue of having CpG sites in their recognition sequence. After digestion, 10 µL of each product were directly loaded onto 2% agarose or 6% polyacrylamide gels and stained with ethidium bromide. Primer sequences and annealing temperatures are shown in Table 1.
Cases and controls were described according to their basic sociodemographic, and clinicopathological factors. Patients who indicated that they had stopped smoking or drinking alcohol within the post 6 months were classified as current smokers or current alcohol drinkers. χ2 tests and Fisher's exact tests were done using the software package SAS Release 8.01 for Windows to examine the differences in DNA methylation status. The corresponding tests on the cases and controls were carried out (p-values) to compare each factor.
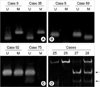
We determined aberrant DNA methylation of hMLH1, MGMT, MINT 25, and GSTP1 in 100 gastric cancer patients and 238 randomly chosen, healthy Koreans. The mean ages were 62 yr for cancer patients and 60.7 yr for controls. These genes were generally unmethylated in the control group, in particular, MINT 25 and GSTP1 showed no hypermethylation. All these genes, however, were aberrantly methylated in the cancer group at the following frequencies: 26 (26%) for hMLH1, 25 (25%) for MGMT, 19 (19%) for MINT 25, and 7 (7%) for GSTP1. All the p values are <0.0001, and the overall results are shown in Fig. 1 and Table 2. When we compared the relationship of the DNA methylation of hMLH1 and other 3 genes, we found concurrent methylation of hMLH1 and any of other 3 genes (p=0.045) (Table 3).
We analyzed the methylation changes in the tumors and the questionnaire data obtained from the patients. The promoter hypermethylation of hMLH1, MGMT, and GSTP1 were detected more frequently in women than in men and the frequencies are as follows: 44.4% vs. 15.6% for hMLH1 (p=0.008), 50.0% vs. 10.9% for MGMT (0.0001), and 13.9% vs. 3.1% for GSTP1 (p=0.0003). To determine the relationship between DNA hypermethylation and aging, we ranked the 100 gastric carcinomas into three groups according to age. The proportion of MINT 25 methylation was increased with age (p=0.037). We compared the frequency of methylation after grouping the cases in two by tobacco smoking or never-smoking. It is interesting to note that the promoter hypermethylation of MGMT and MINT 25 were detected less frequently in the alcohol consumption subgroup and the frequencies are as follows: 14.6% vs. 32.2% for MGMT (p=0.046), and 9.8% vs. 25.4% for MINT 25 (p=0.049). When we compared the methylation after grouping in two by smoking or never smoking, we observed similar results in alcohol consumption, which showed a lower proportion hMLH1 methylation (18.6% vs. 36.6%, p=0.044), MGMT (11.9% vs. 43.9%, p=0.0003), and MINT 25 (11.9% vs. 29.3%, p=0.029). There was no significant difference in the methylation frequency between subgroups with and without gastric cancer family history. The proportion of hMLH1 methylation in these two groups was 27.6% and 15.4%, respectively, but it is not statistically significant.
Another aim of this study was to investigate that the instances of promoter hypermethylation in gastric carcinoma are associated with histological parameters such as H. pylori infection, tumor location, and Lauren classification. In our study, MGMT showed hypermethylation more frequently in distal gastric carcinomas than in the proximal type (34% vs. 16%, p=0.038). These were summarized in Table 4.
Methylation of the CpG islands of tumor suppressor genes leading to their transcriptional inactivation is a highly consistent feature of tumorigenesis. In the present study, we demonstrated the distribution pattern of the aberrant methylation of hMLH1, MGMT, MINT 25 and GSTP1 in gastric cancer patients and controls. We also investigated the association between promoter hypermethylation and relevant parameters including age, gender, alcohol consumption, smoking, and family history. Other histological characteristics were also taken into consideration.
The transcriptional inactivation of MGMT by DNA methylation occurs in a wide spectrum of human tumors (10), whereas that of hMLH1 is restricted to sporadic tumors with microsatellite instability such as colon (11), endometrial (12, 13), and gastric tumors (14). MGMT plays a major role in the repair of O6-methylguanine DNA adducts. The loss of MGMT expression is rarely due to genetic mutation, but due to the methylation of discrete regions of the CpG island of the gene. Recently reported data indicated that MGMT protein expression levels were decreased by the promoter hypermethylation of MGMT in gastric carcinomas (15). Glutathione S-transferases (GSTs) are a family of enzymes involved in the detoxication of xenobiotics and oxygen radicals (16, 17). Recent studies have demonstrated that the expression of the GSTP1 gene, one of the GST isoenzymes, is controlled by DNA methylation (18). MINT 25 stands out as the specific methylation pattern in gastric tumors, and there was no methylation observed
in either normal stomach or colon, or less than 10% of colorectal tumors (19). This suggests that it may play a special role in stomach neoplasia.
In the present study, the methylation of hMLH1, MGMT, MINT 25, and GSTP1 in gastric cancer was detected more frequently than in the controls (p<0.0001). Previous report showed that concurrent hypermethylation of hMLH1, CDH1, MGMT and COX2 gene promoters was more frequently observed in MSI-H gastric tumors, and the significant association between the concurrent hypermethylation and MSI-H was lost when hMLH1 was excluded (20). We tried to compare the hypermethylation pattern between hMLH1 and other genes analyzed in this study. Our result indicates that concurrent hypermethylation of hMLH1 and the other three genes are a common event in gastric cancer (p=0.045). Further studies are necessary to determine the association between the inactivation of hMLH1 gene promoters by hypermethylation and the microsatellite instability status of gastric carcinomas.
We investigated the association between promoter hypermethylation and relevant parameters including age, gender, alcohol consumption, smoking, and family history. We found that the promoter hypermethylation of hMLH1, MGMT, and GSTP1 was detected more frequently in women than in men (p<0.05). A previous report showed that the methylation of TIMP-3 was seen more frequently in women with lung cancer, whereas methylation of DAPK and p16INK4a was more common in men. These data suggest that some genes showed gender-specific methylation pattern, but the reason for this is unknown.
It has been described that aging is associated with the methylation of certain genes such as hMLH1 (21) and estrogen receptor (22). In the present study the proportion of MINT 25 methylation increased with age (p=0.037), and hMLH1 also showed significant difference of methylation frequency between the two age groups of 69 yr apart. Previous studies showed that age-related methylation affects only a subset of genes, suggesting a gene-specific susceptibility in this process (23). Furthermore, there are significant tissue-specific differences in age-related methylation (23). MINT 25, which is strongly associated with gastric carcinoma, demonstrated for the first time in this report an age dependent methylation pattern.
In the present study, there was a significant association between methylation and smoking/alcohol consumption. The proportion of promoter methylation of genes increased significantly in the never-smoking or never-drinking subgroups (p<0.05). It was also shown that the incidence of MGMT promoter hypermethylation was significantly higher in never-smokers in lung adenocarcinomas (24). These result are, however, inconsistent with the previous study related with the promoter methylation pattern of tumor suppressor genes in head and neck squamous cell carcinoma (25). It could be suggested that hypermethylation is regulated differently by alcohol consumption and/or smoking in genes.
There was a difference in hMLH1 methylation between subgroups with and without family history of gastric carcinoma, but this is not statistically significant. Since the size of these two subgroups is significantly different, it needs further investigation with more subgroups without gastric cancer family history.
A previous study showed that the p16 methylation in the distal stomach epithelium was higher than that in the proximal stomach (26). There have been reports showing that the microsatellite instability phenotype is linked with promoter hypermethylation of hMLH1 and MGMT (27). Furthermore, many studies including with Korean patients showed that gastric cancer with microsatellite instability was associated with distal location (28-31). These reports suggest that hypermethylation is more susceptible in distal gastric carcinoma. The correlation mechanism between the microsatellite instability and DNA methylation needs to be uncovered. In our study, MGMT hypermethylation was detected more frequently in distal gastric carcinoma than in the proximal type (p=0.038).
A previous report suggested that there was a significant association between hMLH1 promoter hypermethylation and intestinal type gastric carcinomas (32). Another study, however, showed that the frequency of hMLH1 methylation was similar between intestinal and diffuse type gastric carcinomas (3). In our study, there was no association between hMLH1 hypermethylation and the Lauren classification of gastric carcinomas.
In conclusion, our study regarding promoter hypermethylation of genes involved in the cell repair system and MINT 25 in primary gastric carcinomas shows the high frequency of methylation of hMLH1, MGMT, and MINT 25. This study also demonstrates that hypermethylation was strongly associated with females, and non-drinking/non-smoking subgroups. Moreover, the frequency of MINT 25 hypermethylation increased with aging, and the methylation of MGMT was frequently detected in distal gastric cancer than in proximal cancer. The exact nature of the methylation defect in cancer cells should be defined by further studies.
Figures and Tables
Fig. 1
Methylation analysis in gastric cancer. (A) hMLH1, (B) MGMT, and (C) GSTP1 methylation were analyzed by methylation-specific PCR. The presence of visible PCR products in those lanes marked U indicate the presence of unmethylated genes; the presence of products in those lanes marked M indicate the presence of methylated genes. Cases 9, 35, 59, and 92 show methylated and unmethylated bands because of the heterogeneously methylated genes, and cases 5 and 75 do not have methylated genes. (D) MINT 25 methylation analysis was performed by bisulfite-PCR and restriction digestion. Only methylated alleles will be digested by restriction enzymes, and they are indicated by arrows. Case 27 and 28 have homogeneously and, heterogeneously methylated MINT 25 DNA fragments, respectively.
ACKNOWLEDGEMENT
This research was supported by research grants from Catholic University of Daegu in 2002.
References
2. Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001. 61:3225–3229.
3. To KF, Leung WK, Lee TL, Yu J, Tong JH, Chan MW, Ng EK, Chung SC, Sung JJ. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int J Cancer. 2002. 102:623–628.

4. Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998. 95:11891–11896.

5. Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, Baylin SB, Issa JP. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999. 59:5438–5442.
6. Oue N, Oshimo Y, Nakayama H, Ito R, Yoshida K, Matsusaki K, Yasui W. DNA methylation of multiple genes in gastric carcinoma: Association with histological type and CpG island methylator phenotype. Cancer Sci. 2003. 94:901–905.

7. Sambrook J, Fritsch EF, Maniatis T. Sambrook J, Fritsch EF, Maniatis T, editors. Isolation of high-molecular-weight DNA from mammalian cells. Molecular cloning: A laboratory manual. 1989. 2nd eds. Cold spring harbor, New York: Cold Spring Harbor Lab. Press;9.16–9.22.
8. Wang RY, Gehrke CW, Ehrlich M. Comparison of bisulfite modification of 5-methyldeoxycytidine and deoxycytidine residues. Nucleic Acids Res. 1980. 8:4777–4790.

9. Olek A, Oswald J, Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 1996. 24:5064–5066.

10. Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999. 59:793–797.
11. Wheeler JM, Beck NE, Kim HC, Tomlinson IP, Mortensen NJ, Bodmer WF. Mechanisms of inactivation of mismatch repair genes in human colorectal cancer cell lines: the predominant role of hMLH1. Proc Natl Acad Sci USA. 1999. 96:10296–10301.

12. Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998. 17:2413–2417.

13. Esteller M, Catasus L, Matias-Guiu X, Mutter GL, Prat J, Baylin SB, Herman JG. hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am J Pathol. 1999. 155:1767–1772.

14. Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, Zou TT, Abraham JM, Kong D, Smolinski KN, Shi YQ, Rhyu MG, Powell SM, James SP, Wilson KT, Herman JG, Meltzer SJ. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999. 59:1090–1095.
15. Oue N, Shigeishi H, Kuniyasu H, Yokozaki H, Kuraoka K, Ito R, Yasui W. Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer. 2001. 93:805–809.

16. Pickett CB, Lu AY. Glutathione S-transferases: gene structure, regulation, and biological function. Annu Rev Biochem. 1989. 58:743–764.

17. Tsuchida S, Sato K. Glutathione transferases and cancer. Crit Rev Biochem Mol Biol. 1992. 27:337–384.

18. Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova GS, Hsieh WS, Isaacs WB, Nelson WG. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci USA. 1994. 91:11733–11737.

19. Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999. 96:8681–8686.

20. Carvalho B, Pinto M, Cirnes L, Oliveira C, Machado JC, Suriano G, Hamelin R, Carneiro F, Seruca R. Concurrent hypermethylation of gene promoters is associated with a MSI-H phenotype and diploidy in gastric carcinomas. Eur J Cancer. 2003. 39:1222–1227.

21. Nakajima T, Akiyama Y, Shiraishi J, Arai T, Yanagisawa Y, Ara M, Fukuda Y, Sawabe M, Saitoh K, Kamiyama R, Hirokawa K, Yuasa Y. Age-related hypermethylation of the hMLH1 promoter in gastric cancers. Int J Cancer. 2001. 94:208–211.
22. Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998. 72:141–196.
23. Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998. 58:5489–5494.
24. Pulling LC, Divine KK, Klinge DM, Gilliland FD, Kang T, Schwartz AG, Bocklage TJ, Belinsky SA. Promoter hypermethylation of the O6-methylguanine-DNA methyltransferase gene: more common in lung adenocarcinomas from never-smokers than smokers and associated with tumor progression. Cancer Res. 2003. 63:4842–4848.
25. Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene. 2002. 21:4231–4236.

26. Bai H, Gu L, Zhou J, Deng D. p16 hypermethylation during gastric carcinogenesis of Wistar rats by N-methyl-N-nitro-N-nitrosoguanidine. Mutat Res. 2003. 535:73–78.

27. Musulen E, Moreno V, Reyes G, Sancho FJ, Peinado MA, Esteller M, Herman JG, Combalia N, Rey M, Capella G. Standardized approach for microsatellite instability detection in gastric carcinomas. Hum Pathol. 2004. 35:335–342.
28. Ottini L, Palli D, Falchetti M, D'Amico C, Amorosi A, Saieva C, Calzolari A, Cimoli F, Tatarelli C, De Marchis L, Masala G, Mariani-Costantini R, Cama A. Microsatellite instability in gastric cancer is associated with tumor location and family history in a high-risk population from Tuscany. Cancer Res. 1997. 57:4523–4529.
29. Wu MS, Lee CW, Shun CT, Wang HP, Lee WJ, Sheu JC, Lin JT. Clinicopathological significance of altered loci of replication error and microsatellite instability-associated mutations in gastric cancer. Cancer Res. 1998. 58:1494–1497.
30. Yamamoto H, Perez-Piteira J, Yoshida T, Terada M, Itoh F, Imai K, Perucho M. Gastric cancers of the microsatellite mutator phenotype display characteristic genetic and clinical features. Gastroenterology. 1999. 116:1348–1357.
31. Kim H, Kim YH, Kim SE, Kim NG, Noh SH, Kim H. Concerted promoter hypermethylation of hMLH1, p16INK4A, and E-cadherin in gastric carcinomas with microsatellite instability. J Pathol. 2003. 200:23–31.
32. Oue N, Sentani K, Yokozaki H, Kitadai Y, Ito R, Yasui W. Promoter methylation status of the DNA repair genes hMLH1 and MGMT in gastric carcinoma and metaplastic mucosa. Pathobiology. 2001. 69:143–149.




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download