Abstract
Rapid prenatal diagnosis of common chromosome aneuploidies have been successful through quantitative fluoresent PCR (QF-PCR) assays and small tandem repeat (STR) markers. The purpose of our study was to investigate the clinical feasibility for rapid prenatal detection of Down syndrome using the quantitative fluorescent PCR in uncultured amniocytes. DNA was extracted from uncultured amniotic fluid of normal karyotype (n=200) and of Down syndrome (n=21). It was amplified using QF-PCR with four STR markers located on chromosome 21. Among normal samples, the ranges of diallelic peaks for at least one STR marker were 1.0-1.3 for D21S11, 1.0-1.4 for D21S1411 and 1.0-1.5 for D21S1270. Down syndrome samples showed trisomic triallelic patterns or trisomic diallelic patterns. The sensitivity, specificity, and efficiency of the assay for detecting Down syndrome were 95.4%, 100%, and 99.5%, respectively. Rapid prenatal diagnosis of Down syndrome using QF-PCR is a reliable technique that aids clinical management of pregnancy.
Analysis of uncultured fetal cells by fluorescence in situ hybridization (FISH) has been used as a rapid alternative in screening for common chromosomal aneuploidies (1, 2). Although FISH usually takes only 24 to 48 hr, it is expensive and intensively laborious. Recently, quantitative fluorescent PCR (QF-PCR) of single tandem repeats (STRs) has been developed for rapid prenatal diagnosis of aneuploidies (3, 4). This approach has been used extensively by several groups on research basis (5-10), and the diagnosis of aneuploidies by QF-PCR of STRs has now been validated as a reliable method applicable in many laboratories.
In this study, we investigated the clinical feasibility of QF-PCR for rapid prenatal diagnosis of trisomy 21.
Genomic DNA extraction was performed on the cell pellet obtained from 1 to 2 mL of amniotic fluid from 200 normal samples and 21 samples with trisomy 21. The karyotypes of all amniotic fluid samples were performed previously by conventional cytogenetic analysis. DNA was extracted by incubating cell pellets with InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA, U.S.A.). A single tube multiplex PCR was carried out by STR markers specific for chromosome 21. D21S11, D21S1411, and D21S1270 (10) were used to test all samples for chromosome 21. D21S1412 (11) was added to test samples found to be homozygous for the first three STR (Table 1). The QF-PCR amplification of STR markers was performed in a total volume of 25 µL containing 1.5 mM MgCl2, 200 µm/L dNTP, 10-30 pmol of each primer, PCR buffer, 2U Taq polymerase, and 10 µL genomic DNA. After the initial denaturation at 94℃ for 5 min, hot start PCR was followed by 25 cycles of 94℃ for 35 sec, 58℃ for 35 sec, 72℃ for 40 sec and final extension was for 5 min at 72℃. For fragment analysis of PCR products, ABI 3100 and GeneScan 3.7 (Applied Biosystems, Foster City, CA, U.S.A.) were used.
The statistical analysis of the heterozygosity for each STR marker between the published population (10) and the Korean population were analyzed using the Z-test. A p-value >1.96 was taken as significant.
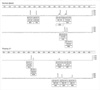
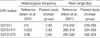
The majority of normal samples showed diallelic peaks with a ratio of 1:1 for each STR marker (Fig. 1). Among normal samples, the ranges of diallelic peaks were 1.0-1.3 for D21S11, 1.0-1.4 for D21S1411 and 1.0-1.5 for D21S1270 (Table 2). Down syndrome samples that confirmed karyotypes with 47,+21 showed trisomic triallelic patterns with an allele peak ratio 1:1:1, or trisomic diallelic patterns with an allele peak ratio 2:1 (Fig. 1). However, one case that showed low-level mosaicism with a karyotype of 47,XX,+21[23]/46,XX [218] failed to be detected by QF-PCR. In complete trisomic 21 samples, the ranges of diallelic peaks were 1.6-2.4 for D21S11, 2.2-2.7 for D21S1411, and 1.7-2.9 for D21S1270 (Table 3).
All but one normal samples were heterozygous and informative for at least one STR marker (Table 4).
The sensitivity, specificity, and efficiency of QF-PCR for the detection of Down syndrome were 95.4%, 100%, and 99.5%, respectively. The false positive and the false negative rates were 0% (0/200) and 4.7% (1/21), respectively. All results were obtained within 24 to 48 hr.
QF-PCR for the detection of chromosome specific repeated sequences has been improved to include several STR (3, 4). It is based on the incorporation of fluorochromes into the products of PCR amplification via oligonucleotide primers specific for each STR and on the assumption that, within the early exponential phase of PCR amplification, the amount of specific STR produced is proportional to the quantity of initial target sequence (12). In normal heterozygotes, the ratio of the fluorescent intensity of the two peaks should be close to 1:1. If the STR marker is highly polymorphic, few normal subjects should be homozygotes and show one peak. In a trisomic patient, the three doses of an STR marker can be detected either as three peaks of fluorescent intensity with a ratio 1:1:1 (trisomic triallelic) or as a pattern of two peaks with a ratio 2:1 (trisomic diallelic). Using the high polymorphism of the STR markers, very few trisomic patients should show a single peak of fluorescent intensity (4).
Our results present that the fluorescent intensity ratios showed a definite difference between normal disomic diallelic samples and diallelic trisomy 21 samples.
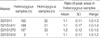
In the present study, we used the markers as published previously. It was shown that heterozygosity of markers was low in our population compared to the published data (Table 5). At the D21S11 and D21S1411 markers, a significant differences were found. We expected a difference of genetic diversity between an ethnic groups and it was supported by other previous reports (13-16).
Although 4 samples showed heterozygous for only one STR, even after using the extra marker, the use of combined STR markers could reduce the likelihood of homozygosity and consequently, the frequency of uninformative STR patterns. Only one out of 220 samples was homozygous for all STR tested (Table 4). As a results, we emphasize that the informative rate of our experiment is prominent than other studies so far (5, 8, 9, 16).
Yoon et al. reported three false positive in the multiplex QF-PCR using two of STR markers for Down syndrome (16). As it is well known, the preferential amplification of multiplex PCR is a potential problem that result in an incorrect genetic typing and is effected by low denaturation temperatures, low amount of DNA, and differential allelic priming. In this study, there were no false positive results from multiplex QF-PCR using three of STR markers. It is reported that a maximum primer set used in the multiplex QF-PCR is twelve up to now (10).
One case of low level mosaicism for trisomy 21 with a karyotype of 47,XX,+21[23]/46,XX[218] resulted in a false negative result. Several studies reported that mosaicism for trisomy was able to be detected by QF-PCR (10, 11), but based on our own and other published experiences, mosaicism, maternal contamination, structural abnormality, deletion and duplication syndrome are not likely to be detected by the QF-PCR technique (8, 9). However, it is possible that future development, such as the use of extensive panel of markers to cover other key chromosomes, will result in significant improvements in the detection rate.
An advantage of using QF-PCR technique is that it is less time consuming and less laborious and this assay is useful on small volumes of amniotic fluid (1-2 mL). In this study, some results were obtained within 8 hr.
Our experiment demonstrates that QF-PCR can provide a rapid and accurate clinical method for prenatal identification of Down syndrome and also serve as an adjunctive test to help cytogenetics to reduce significant amounts of emotional stress experienced by patients and physicians. In further studies, we will test a larger scale population with expanded panel of markers for other numerical aberrations, such as trisomy 13 or 18 and sex chromosome aneuploidies.
Figures and Tables
Fig. 1
Electrophoregram of the QF-PCR products from normal and trisomy 21 samples. Fragment size in bp shown on horizontal axis, arbitrary fluorescence units shown on vertical axis. Markers labelled according to locus, size, and peak areas. Normal diploid sample: all markers are heterozygous and exhibit two peaks with a 1:1 ratio. Trisomy 21 sample: chromosome 21 markers exhibit three peaks with a 1:1:1 ratio or two peaks with a 2:1 ratio.
References
1. Klinger K, Landes G, Shook D, Harvey R, Lopez L, Locke P, Lerner T, Osathanondh R, Leverone B, Houseal T. Rapid detection of chromosome aneuploidies in uncultured amniocytes by using fluorescence in situ hybridization (FISH). Am J Hum Genet. 1992. 51:55–65.
2. Thilaganathan B, Sairam S, Ballard T, Peterson C, Meredith R. Effectiveness of prenatal chromosomal analysis using multicolor fluorescent in situ hybridization. Bjog. 2000. 107:262–266.
3. Mansfield ES. Diagnosis of Down syndrome and other aneuploidies using quantitative polymerase chain reaction and small tandem repeat polymorphisms. Hum Mol Genet. 1993. 2:43–50.

4. Pertl B, Yau SC, Sherlock J, Davies AF, Mathew CG, Adinolfi M. Rapid molecular method for prenatal detection of Down's syndrome. Lancet. 1994. 343:1197–1198.

5. Pertl B, Weitgasser U, Kopp S, Kroisel PM, Sherlock J, Adinolfi M. Rapid detection of trisomies 21 and 18 and sexing by quantitative fluorescent multiplex PCR. Hum Genet. 1996. 98:55–59.

6. Toth T, Findlay I, Papp C, Toth-Pal E, Marton T, Nagy B, Quirke P, Papp Z. Prenatal detection of trisomy 21 and 18 from amniotic fluid by quantitative fluorescent polymerase chain reaction. J Med Genet. 1998. 35:126–129.

7. Verma L, Macdonald F, Leedham P, McConachie M, Dhanjal S, Hulten M. Rapid and simple prenatal DNA diagnosis of Down's syndrome. Lancet. 1998. 352:9–12.

8. Schmidt W, Jenderny J, Hecher K, Hackeloer BJ, Kerber S, Kochhan L, Held KR. Detection of aneuploidy in chromosomes X, Y, 13, 18 and 21 by QF-PCR in 662 selected pregnancies at risk. Mol Hum Reprod. 2000. 6:855–860.

9. Levett LJ, Liddle S, Meredith R. A large-scale evaluation of amnio-PCR for the rapid prenatal diagnosis of fetal trisomy. Ultrasound Obstet Gynecol. 2001. 17:115–118.

10. Mann K, Fox SP, Abbs SJ, Yau SC, Scriven PN, Docherty Z, Ogilvie CM. Development and implementation of a new rapid aneuploidy diagnostic service within the UK National Health Service and implications for the future of prenatal diagnosis. Lancet. 2001. 358:1057–1061.

11. Pertl B, Kopp S, Kroisel PM, Hausler M, Sherlock J, Winter R, AdinolfiM . Quantitative fluorescence polymerase chain reaction for the rapid prenatal detection of common aneuploidies and fetal sex. Am J Obstet Gynecol. 1997. 177:899–906.

12. Ferre F. Quantitative or semi-quantitative PCR: reality versus myth. PCR Methods Appl. 1992. 2:1–9.

13. Han GR, Lee YW, Lee HL, Kim SM, Ku TW, Kang IH, Lee HS, Hwang JJ. A Korean population study of the nine STR loci FGA, VWA, D3S1358, D18S51, D21S11, D8S1179, D7S820, D13S317 and D5S818. Int J Legal Med. 2000. 114:41–44.

14. Lee JW, Lee HS, Hwang JJ. Statistical analysis for estimating heterogeneity of the Korean population in DNA typing using STR loci. Int J Legal Med. 2002. 116:153–160.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download