Abstract
Background:
Clostridium difficile is a major cause of antibiotic-associated diarrhea. The objective of this study was to characterize clinical isolates of C. difficile obtained from various regions in Korea with regard to their toxin status, molecular type, and antimicrobial susceptibility.
Methods:
We analyzed a total of 408 C. difficile isolates obtained between 2006 and 2008 from 408 patients with diarrhea in 12 South Korean teaching hospitals. C. difficile toxin genes tcdA, tcdB, cdtA, and cdtB were detected by PCR. Molecular genotyping was performed by PCR ribotyping. Antimicrobial susceptibilities of the 120 C. difficile isolates were assessed by agar dilution methods.
Results:
Among 337 toxigenic isolates, 105 were toxin A-negative and toxin B-positive (A-B+) and 29 were binary toxin-producing strains. PCR ribotyping showed 50 different ribotype patterns. The 5 most frequently occurring ribotypes comprised 62.0% of all identified ribotypes. No isolate was susceptible to cefoxitin, and all except 1 were susceptible to piperacillin and piperacillin-tazobactam. The resistance rates of isolates to imipenem, cefotetan, moxifloxacin, ampicillin, and clindamycin were 25%, 34%, 42%, 51%, and 60%, respectively. The isolates showed no resistance to metronidazole or vancomycin.
Conclusions:
This is the first nationwide study on the toxin status, including PCR ribotyping and antimicrobial resistance, of C. difficile isolates in Korea. The prevalence of A-B+ strains was 25.7%, much higher than that reported from other countries. Binary toxin-producing strains accounted for 7.1% of all strains, which was not rare in Korea. The most prevalent ribotype was ribotype 017, and all A-B+ strains showed this pattern. We did not isolate strains with decreased susceptibility to metronidazole or vancomycin.
REFERENCES
1.Bartlett JG. Clostridium difficile: history of its role as an enteric pathogen and the current state of knowledge about the organism. Clin Infect Dis. 1994. 18(S4):265–72.
2.Borriello SP., Davies HA., Kamiya S., Reed PJ., Seddon S. Virulence factors of Clostridium difficile. Rev Infect Dis. 1990. 12(S2):185–91.
3.Braun V., Hundsberger T., Leukel P., Sauerborn M., von Eichel-Streiber C. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene. 1996. 181:29–38.
4.Hammond GA., Johnson JL. The toxigenic element of Clostridium difficile strain VPI 10463. Microb Pathog. 1995. 19:203–13.
5.Borriello SP., Wren BW., Hyde S., Seddon SV., Sibbons P., Krishna MM, et al. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect Immun. 1992. 60:4192–9.
6.Lyerly DM., Barroso LA., Wilkins TD., Depitre C., Corthier G. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect Immun. 1992. 60:4633–9.
7.Alfa MJ., Kabani A., Lyerly D., Moncrief S., Neville LM., Al-Barrak A, et al. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J Clin Microbiol. 2000. 38:2706–14.
8.Popoff MR., Rubin EJ., Gill DM., Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun. 1988. 56:2299–306.
9.Perelle S., Gibert M., Bourlioux P., Corthier G., Popoff MR. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect Immun. 1997. 65:1402–7.
10.Barbut F., Mastrantonio P., Delmée M., Brazier J., Kuijper E., Poxton I. Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin Microbiol Infect. 2007. 13:1048–57.
11.McDonald LC., Killgore GE., Thompson A., Owens RC Jr., Kazakova SV., Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005. 353:2433–41.
12.Warny M., Pepin J., Fang A., Killgore G., Thompson A., Brazier J, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005. 366:1079–84.
13.Baines SD., O'Connor R., Freeman J., Fawley WN., Harmanus C., Mastrantonio P, et al. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J Antimicrob Chemother. 2008. 62:1046–52.
14.Muto CA., Pokrywka M., Shutt K., Mendelsohn AB., Nouri K., Posey K, et al. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect Control Hosp Epidemiol. 2005. 26:273–80.
15.Kato H., Kato N., Watanabe K., Iwai N., Nakamura H., Yamamoto T, et al. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998. 36:2178–82.
16.Stubbs S., Rupnik M., Gibert M., Brazier J., Duerden B., Popoff M. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000. 186:307–12.
17.O'Neill GL., Ogunsola FT., Brazier JS., Duerden BI. Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe. 1996. 2:205–9.
18.Spigaglia P., Mastrantonio P. Comparative analysis of Clostridium difficile clinical isolates belonging to different genetic lineages and time periods. J Med Microbiol. 2004. 53:1129–36.
19.Clinical and Laboratory Standards Institute. Methods for antimicrobial susceptibility testing of Anaerobic bacteria; Approved standard. 7th ed.CLSI document M11-A7. Wayne, PA: Clinical and Laboratory Standards Institute;2007.
20.Huang H., Fang H., Weintraub A., Nord CE. Distinct ribotypes and rates of antimicrobial drug resistance in Clostridium difficile from Shanghai and Stockholm. Clin Microbiol Infect. 2009. 15:1170–3.
21.Kim H., Riley TV., Kim M., Kim CK., Yong D., Lee K, et al. Increasing prevalence of toxin A-negative, toxin B-positive isolates of Clostridium difficile in Korea: impact on laboratory diagnosis. J Clin Microbiol. 2008. 46:1116–7.
22.Shin BM., Kuak EY., Yoo HM., Kim EC., Lee K., Kang JO, et al. Multi-centre study of the prevalence of toxigenic Clostridium difficile in Korea: results of a retrospective study 2000-2005. J Med Microbiol. 2008. 57:697–701.
23.Geric B., Rupnik M., Gerding DN., Grabnar M., Johnson S. Distribution of Clostridium difficile variant toxinotypes and strains with binary toxin genes among clinical isolates in an American hospital. J Med Microbiol. 2004. 53:887–94.
24.Martin H., Willey B., Low DE., Staempfli HR., McGeer A., Boerlin P, et al. Characterization of Clostridium difficile strains isolated from patients in Ontario, Canada, from 2004 to 2006. J Clin Microbiol. 2008. 46:2999–3004.
25.Keel K., Brazier JS., Post KW., Weese S., Songer JG. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J Clin Microbiol. 2007. 45:1963–4.
26.Goorhuis A., Bakker D., Corver J., Debast SB., Harmanus C., Notermans DW, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008. 47:1162–70.
27.Huang H., Weintraub A., Fang H., Nord CE. Antimicrobial resistance in Clostridium difficile. Int J Antimicrob Agents. 2009. 34:516–22.
28.Drudy D., Quinn T., O'Mahony R., Kyne L., O'Gaora P., Fanning S. High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J Antimicrob Chemother. 2006. 58:1264–7.
29.John R., Brazier JS. Antimicrobial susceptibility of polymerase chain reaction ribotypes of Clostridium difficile commonly isolated from symptomatic hospital patients in the UK. J Hosp Infect. 2005. 61:11–4.
Fig. 1.
PCR ribotype patterns of the Clostridium difficile isolates representing PCR ribotypes AB24, C11, C5, C2, AB14, AB23, and aB (Lane 1 to 7, respectively). Lane L refers to 100 bp ladder. Banding patterns of the C5, C2, and aB ribotypes were identical to the pattern of C. difficile ribotype 027, 078, and 017 strains.
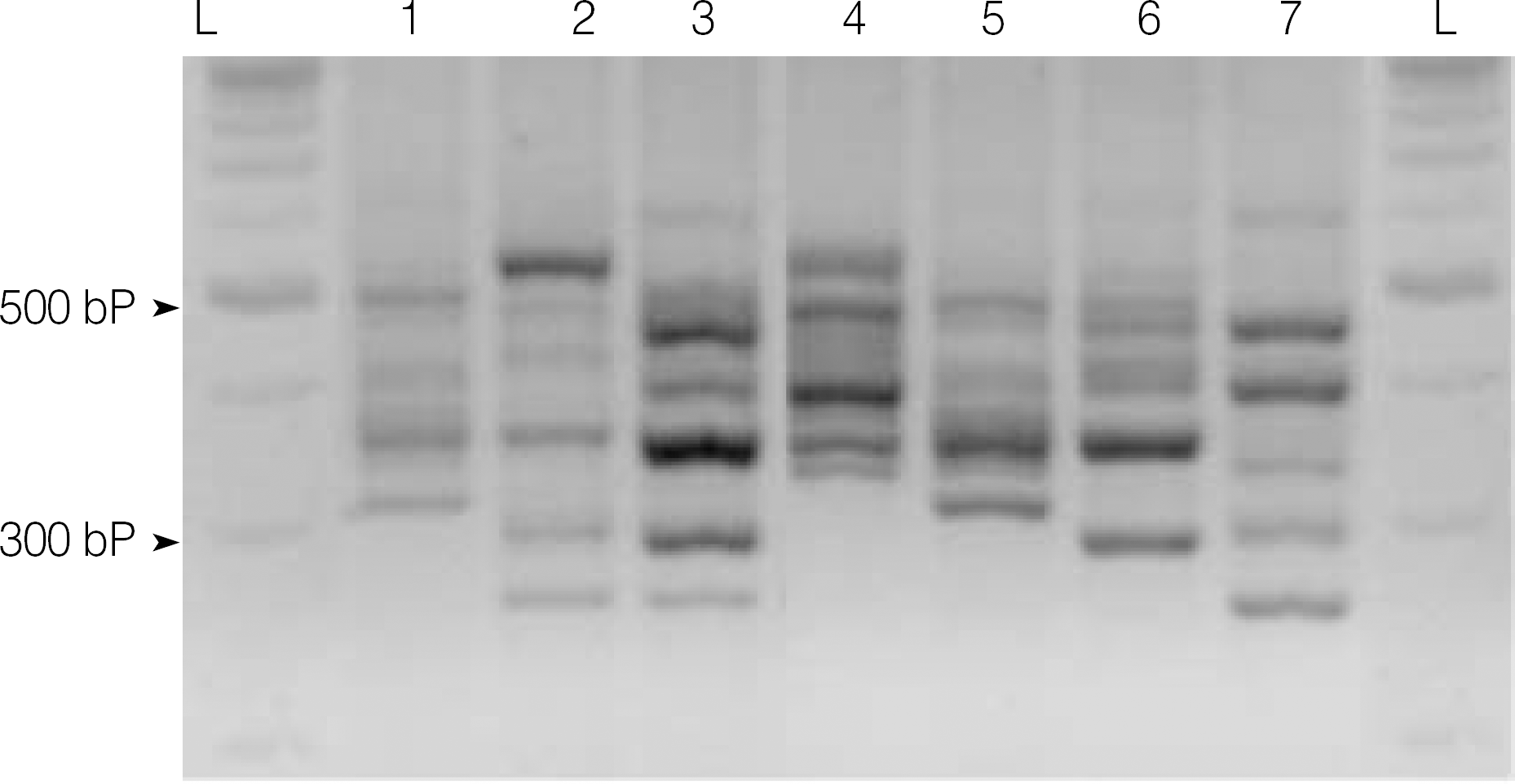
Table 1.
The frequencies of the occurrences of toxins A-, B-, and binary toxin-producing strains in 12 South Korean hospitals
| Hospitals | N of isolates tested | Study period | Number of beds∗ | N (%) | |||
|---|---|---|---|---|---|---|---|
| A+B+CDT- | A+B+CDT+ | A-B+CDT- | A-B-CDT- | ||||
| Seoul A | 145 | Jan.2007-Dec. 2007 | 2,064 | 52 (35.9) | 8 (5.5) | 55 (37.9) | 30 (20.7) |
| Seoul B | 37 | Jan.2007-Jun.2008 | 758 | 19 (51.4) | 4 (10.8) | 9 (24.3) | 5 (13.5) |
| Seoul C | 30 | Jan.2007-Feb.2008 | 938 | 13 (43.3) | 6 (20.0) | 6 (20.0) | 5 (16.7) |
| Seoul D | 20 | Jan.2008-Feb.2008 | 2,200 | 8 (40.0) | 3 (15.0) | 3 (15.0) | 6 (30.0) |
| Gyeonggi A | 41 | Jun.2007-Mar.2008 | 589 | 23 (56.1) | 2 (4.9) | 9 (21.9) | 7 (17.1) |
| Gyeonggi B | 17 | Jan. 2008-May. 2008 | 920 | 14 (82.3) | 1 (5.9) | 2 (11.8) | 0 (0) |
| Gyeonggi C | 15 | Jan.2006-Dec. 2006 | 550 | 8 (53.3) | 0 (0) | 3 (20.0) | 4 (26.7) |
| Chungnam | 22 | Oct.2007-May.2008 | 803 | 12 (54.5) | 2 (9.1) | 6 (27.3) | 2 (9.1) |
| Daejeon | 25 | Mar.2008-Jun.2008 | 813 | 17 (68.0) | 2 (8.0) | 3 (12.0) | 3 (12.0) |
| Busan | 20 | Feb.2007-Dec. 2007 | 912 | 12 (60.0) | 0 (0) | 6 (30.0) | 2 (10.0) |
| Gwangju | 20 | Mar.2008-Jun.-2008 | 555 | 15 (75.0) | 0 (0) | 0 (0) | 5 (25.0) |
| Gangwon | 16 | Nov.2007-May.2008 | 816 | 10 (62.5) | 1 (6.3) | 3 (18.8) | 2 (12.5) |
| Total | 408 | 203 (56.9) | 29 (7.1) | 105 (25.7) | 71 (17.4) | ||
Table 2.
The MICs of 10 antimicrobial agents for 120 Korean Clostridium difficile isolates




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download