Abstract
Background:
Rapid identification of the causative agent among potential bacterial and viral pathogens is important for the management of acute respiratory disease. In this study, we evaluated the analytical performance and clinical usefulness of a recently-introduced multiplex PCR assay, Seeplex™ Pneumobacter detection kit (Seegene Inc., Korea) for the identification of respiratory bacterial pathogens.
Methods:
One hundred and eighty one nasopharyngeal aspirates were collected from pediatric patients with respiratory symptoms and analysed by multiplex PCR for the detection of Streptococcus pneumoniae (S.P), Haemophilus influenzae (H.I), Mycoplasma pneumoniae (M.P), Chlamydophila pneumoniae (C.P), Bordetella pertussis (B.P) and Legionella pneumophila (L.P). A comparison of multiplex PCR with conventional culture for the isolation of S.P and H.I was performed on 112 specimens. The cross reactivity of multiplex PCR was also evaluated.
Results:
Of 181 cases, 81 cases were positive by multiplex PCR (44.8%): 52 cases for S.P (28.7%), 47 cases for H.I (26.0%), 9 cases for M.P (5.0%), 3 cases for B.P (1.7%) and 1 case for C.P (0.6%) including multiple infection cases. The agreement rates between multiplex PCR and culture for S.P and H.I were 92.9% (kappa index=0.84, P<0.001) and 91.1% (kappa index=0.75, P<0.001), respectively. There was no cross reactivity with common bacterial and viral pathogens.
Conclusions:
Seeplex™ Pneumobacter detection kit could be a useful screening tool for the rapid detection of respiratory bacterial pathogens. Further studies with lower respiratory tract specimens would be needed for the clinical evaluation of S. pneumoniae and H. influenzae detected by multiplex PCR.
REFERENCES
1.Leowski J. Mortality from acute respiratory infections in children under 5 years of age: global estimates. World Health Stat Q. 1986. 39:138–44.
2.McCracken GH Jr. Diagnosis and management of pneumonia in children. Pediatr Infect Dis J. 2000. 19:924–8.
3.Fine MJ., Smith MA., Carson CA., Mutha SS., Sankey SS., Weissfeld LA, et al. Prognosis and outcomes of patients with community-acquired pneumonia. A meta-analysis. JAMA. 1996. 275:134–41.

4.Ostapchuk M., Roberts DM., Haddy R. Community-acquired pneumonia in infants and children. Am Fam Physician. 2004. 70:899–908.
5.Fedorko DP., Emery DD., Franklin SM., Congdon DD. Evaluation of a rapid enzyme immunoassay system for serologic diagnosis of Mycoplasma pneumoniae infection. Diagn Microbiol Infect Dis. 1995. 23:85–8.
6.Wang Y., Kong F., Yang Y., Gilbert GL. A multiplex PCR-based reverse line blot hybridization (mPCR/RLB) assay for detection of bacterial respiratory pathogens in children with pneumonia. Pediatr Pulmonol. 2008. 43:150–9.

7.Miyashita N., Saito A., Kohno S., Yamaguchi K., Watanabe A., Oda H, et al. Multiplex PCR for the simultaneous detection of Chlamydia pneumoniae, Mycoplasma pneumoniae and Legionella pneumophilia in community-acquired pneumonia. Respir Med. 2004. 98:542–50.
8.Strålin K., Bäckman A., Holmberg H., Fredlund H., Olcén P. Design of a multiplex PCR for Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae and Chlamydophila pneumoniae to be used on sputum samples. APMIS. 2005. 113:99–111.
9.Sung H., Park SJ., Woo YD., Choi BH., Kim MN. Evaluation of Seeplex RV detection kit for detecting rhinovirus, human metapneumovirus, and coronavirus. Korean J Lab Med. 2008. 28:109–17. (성흥섭, 박숙자,우영대, 최병후, 김미나. Rhinovirus, human metapneumovirus, coronavirus 검출을 위한 Seeplex™ RV Detection 키트의 평가 대한진단검 사의학회지 2008;28:109-17.).
10.Kim SR., Ki CS., Lee NY. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system-based multiplex PCR assay. J Virol Methods. 2009. 156:111–6.

11.Waites KB., Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004. 17:697–728.
12.Choi SK., Jung JA., Kim KH., Kim GH. Study of seroprevalence of antimycoplasma antibody in healthy children and its diagnostic value. J Korean Pediatr Soc. 1998. 41:489–97. (최수경, 정지아, 김경효,김경희. 건강한 소아에서 Mycoplasma 항체가의 분포 및 이의 진단적유용성에관한연구. 소아과 1998;41:489-97.).
13.Bae SM., Jang MJ., Song HJ., Jeon DY., Kweon SS., Kang YH. Prevalence of Mycoplasma pneumoniae antibodies in healthy residents of Jeonnam Province. Korean J Clin Microbiol. 2007. 10:109–13. (배송미,장미정, 송현제, 전두영, 권순석, 강연호. 전남지역주민에서마이코플라 스마폐렴에대한항체가분포. 대한임상미생물학회지 2007;10:109-13.).
14.Räty R., Rönkkö E., Kleemola M. Sample type is crucial to the diagnosis of Mycoplasma pneumoniae pneumonia by PCR. J Med Microbiol. 2005. 54:287–91.
15.Kuo CC., Jackson LA., Campbell LA., Grayston JT. Chlamydia pneumoniae (TWAR). Clin Microbiol Rev. 1995. 8:451–61.
16.Benson RF., Fields BS. Classification of the genus Legionella. Semin Respir Infect. 1998. 13:90–9.
17.Boman J., Allard A., Persson K., Lundborg M., Juto P., Wadell G. Rapid diagnosis of respiratory Chlamydia pneumoniae infection by nested touchdown polymerase chain reaction compared with culture and antigen detection by EIA. J Infect Dis. 1997. 175:1523–6.
18.Hindiyeh M., Carroll KC. Laboratory diagnosis of atypical pneumonia. Semin Respir Infect. 2000. 15:101–13.

19.Gray BM., Converse GM 3rd., Dillon HC Jr. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis. 1980. 142:923–33.
20.Mandell LA., Marrie TJ., Grossman RF., Chow AW., Hyland RH. Canadian guidelines for the initial management of community-acquired pneumonia: an evidence-based update by the Canadian Infectious Diseases Society and the Canadian Thoracic Society. The Canadian Community-Acquired Pneumonia Working Group. Clin Infect Dis. 2000. 31:383–421.
21.Menéndez R., Córdoba J., de La Cuadra P., Cremades MJ., López-Hontagas JL., Salavert M, et al. Value of the polymerase chain reaction assay in noninvasive respiratory samples for diagnosis of community-acquired pneumonia. Am J Respir Crit Care Med. 1999. 159:1868–73.

22.Nakayama E., Hasegawa K., Morozumi M., Kobayashi R., Chiba N., Iitsuka T, et al. Rapid optimization of antimicrobial chemotherapy given to pediatric patients with community-acquired pneumonia using PCR techniques with serology and standard culture. J Infect Chemother. 2007. 13:305–13.

Fig. 1.
Multiplex PCR products of six positive samples. M, 100 bp DNA ladder; lane 1, M. pneumoniae (583 bp); lane 2, L. pneumophila (472 bp); lane 3, S. pneumoniae (350 bp); lane 4, H. influenzae (257 bp); lane 5, B. pertussis (200 bp), lane 6, C. pneumoniae (146 bp); P, positive control ladder; N, negative control.
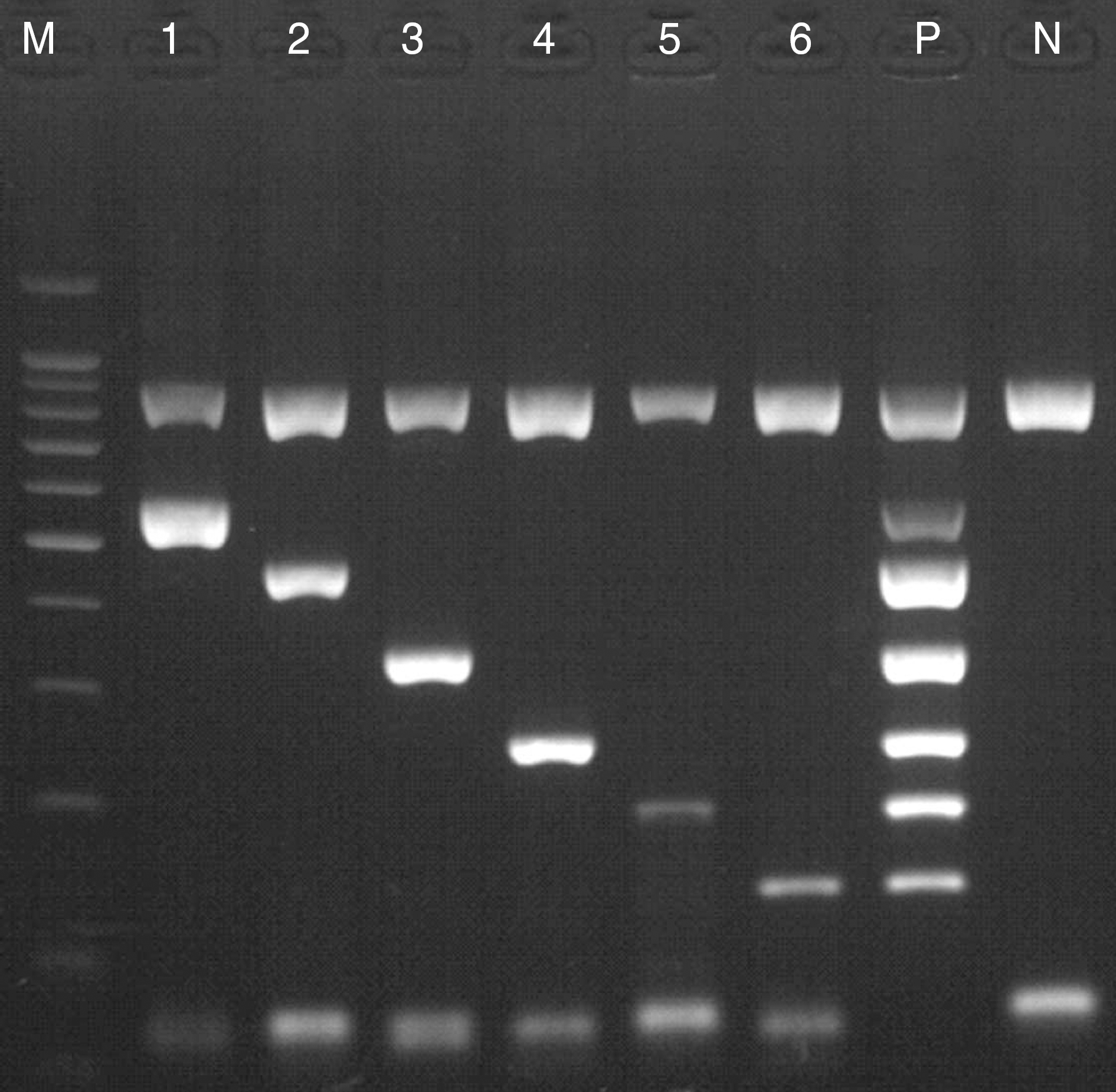
Table 1.
Target genes used in multiplex PCR
Table 2.
Results of analysis on 181 nasopharyngeal aspirates by multiplex PCR
Table 3.
Analytical specificity of multiplex PCR with 21 different microorganisms
Table 4.
Results of multiplex PCR for potential respiratory bacterial and viral pathogens
Table 5.
Comparison of multiplex PCR and conventional culture method for the detection of S. pneumoniae and H. influenzae in 112 patients
Table 6.
Disease entities of positive cases by multiplex PCR




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download