Abstract
MicroRNAs (miRNAs), small noncoding RNAs 19–22 nucleotides in length, play a major role in negative regulation of gene expression at the posttranscriptional level. Several miRNAs act as tumor suppressors or oncogenes that control cell differentiation, proliferation, apoptosis, or angiogenesis during tumorigenesis. To date, 19 research groups have published large-scale expression profiles that identified 261 miRNAs differentially expressed in bladder cancer, of which 76 were confirmed to have consistent expression patterns by two or more groups. These consistently expressed miRNAs participated in regulation of multiple biological processes and factors, including axon guidance, cancer-associated proteoglycans, and the ErbB and transforming growth factorbeta signaling pathways. Because miRNAs can be released from cancer cells into urine via secreted particles, we propose that miRNAs differentially expressed between tissue and urine could serve as predictors of bladder cancer, and could thus be exploited for noninvasive diagnosis.
Bladder cancer is the second most common urological malignancy in humans. In 2012, an estimated 73,510 new cases of bladder cancer were diagnosed in the United States, and 14,680 deaths from bladder cancer were reported [1]. Likewise, in Korea, bladder cancer is the second most common genitourinary tumor, and is about 5 times more common in men than in women [2]. Nonmuscle invasive bladder cancer (NMIBC) is associated with better survival than other malignancies; however, 30%–50% of patients with NMIBC will eventually experience recurrence after transurethral resection of the primary tumor. Moreover, 10%–20% of NMIBC cases progress to muscle invasive bladder cancer (MIBC) [34], as defined by pathology and clinical features [56]. Although only 20% of bladder cancer is confirmed as MIBC at first diagnosis, this subtype accounts for the majority of cancer-specific deaths [7]. Therefore, the major challenge in treating patients with NMIBC is to prevent recurrence and progression to MIBC. Because it is a heterogeneous disease, however, optimal treatment and follow-up strategies vary depending on initial clinical and histopathological characteristics.
A two-pathway model has been proposed for urothelial cell carcinoma pathogenesis. NMIBC recurs in about 70% of cases which harbor mutations in 30%–40% HRAS gene and less than 70% FGFR3 indicating that RTK-Ras activation plays an early and crucial role in this tumorigenic pathway, and is thus a potential prognostic molecular marker [8910]. MIBC accounts for 20%–30% of urothelial tumors with structural and functional defects in TP53, RB, and PTEN. These mutations in turn cause alterations in the tumor microenvironment, including aberrant expression of N- and E-cadherins, matrix metalloproteinases, angiogenic factors such as vascular endothelial growth factor, and antiangiogenic factors such as thrombospondin 1 and cyclooxygenase 2). In addition, over 50% of these tumors progress to local and distant metastases despite radical cystectomy and chemotherapy [8910111213]. Although invasive tumors have larger burdens of mutations of all types, they also exhibit greater clonal diversity, and stage is not clearly correlated with mutation spectrum, suggesting that invasion is not driven by mutagen exposure or specific defects in DNA repair [14]. Nevertheless, a considerable amount of heterogeneity in clinical behavior cannot be explained by a single model.
miRNAs are a class of small (~22 nucleotides) noncoding RNAs that function as negative regulators of gene expression at the post-transcriptional level. They act by binding to complementary sequences in the 3'-untranslated regions of specific mRNAs, resulting in the inhibition of translation [15]. To date, miRNAs have been implicated in the control of many fundamental cellular and physiological processes, including cellular differentiation, proliferation, apoptosis, metabolism, development, and cancer pathogenesis [161718]. Expression of many miRNAs is altered in tumors relative to normal tissues, and such aberrant miRNA expression may be important for tumor progression. The identification of target genes associated with aberrantly expressed miRNAs might elucidate the roles of miRNAs in cancer biology [19]. Earlier studies demonstrated that miRNA expression profiles could discriminate between malignant and nonmalignant tissue, as well as among various tumor entities [2021222324].
Beginning in the 1990s, microarray technology provided a means to simultaneously measure the expression levels of all genes in the genome. In each spot of a microarray, a high concentration of a defined DNA or oligonucleotide is immobilized, and these molecules can undergo specific hybridization with complementary sequences in a sample of interest. A single microarray can contain thousands or millions of specific spots, enabling analysis of the entire transcriptome of a cell at a given time. Microarrays have dramatically accelerated many types of investigation, and have become essential tools for transcriptome analysis.
Several miRNA microarray platforms are commercially available. The GeneChip miRNA 4.0 Array (Affymetrix, Santa Clara, CA, USA) contains spots corresponding to 1,996 human small nucleolar RNAs (snoRNAs), small Cajal body-specific RNAs (scaRNAs), and all pre-miRNA hairpin sequences from miRBase v20. The SurePrint Human miRNA Microarray Slide v21.0 (Agilent, Santa Clara, CA, USA) contains spots corresponding to 2,549 human miRNAs. The 40–60-mer oligonucleotide probes were designed using an optimized method, and are synthesized directly on the array using SurePrint inkjet technology. The Human v2 MicroRNA Expression Profiling Assay (Illumina, San Diego, CA, USA) microarray contains spots corresponding to 1,146 human miRNAs, and can accurately quantitate levels of a diverse population of miRNAs via DASL (cDNA-Mediated Annealing, Selection, Extension, and Ligation) assay.
To date, microarrays remain the most popular approach for transcript profiling, and are affordable for most laboratories. Nonetheless, array technology has several limitations; in particular, it only provides a semiquantitative assessment of gene expression, and background hybridization limits the accuracy of expression measurements, particularly for transcripts present at low levels. Furthermore, probes differ considerably in their hybridization properties, and arrays are limited to interrogating only those genes for which probes can be designed [25].
By contrast, RNA sequencing (RNA-Seq) involves direct sequencing of transcripts by high-throughput sequencing technologies (also called next-generation sequencing, NGS) (Fig. 1). This approach has the potential to replace microarrays for whole-genome transcriptome profiling [26272829]. RNA-Seq has considerable advantages for examination of the fine structure of the transcriptome, including its ability to detect novel transcripts, allele-specific expression, and splice junctions. Moreover, RNA-Seq does not depend on genome annotation for probe selection, and it avoids the associated biases introduced during hybridization of microarrays. However, despite the fact that many computational methods have been developed for read alignment, quantitation of transcripts, and identification of differentially expressed genes [30], RNA-Seq poses novel algorithmic and logistical challenges for data analysis and storage. The available computational tools vary considerably in maturity.
Several studies have compared RNA-Seq and hybridization-based microarrays [313233]. Most RNA-Seq is performed using the Illumina or Ion Torrent platform. Therefore, cross-platform comparisons were conducted in which the same samples were studied using Illumina and Ion Torrent RNA-Seq, as well as hybridization-based approaches. On the Illumina platform, the samples were processed using the TruSeq protocol and sequenced on a HiSeq 2500, yielding 100 × 100-nt paired-end reads. On the Ion Torrent platform, the samples were processed using the Total RNA-Seq V2 protocol, which preserves strand specificity and is capable of capturing non-coding RNA; these libraries were sequenced on an Ion Proton P1 chip, yielding up to 200-nt reads. The data obtained using both platforms were compared in regard to quality, alignment statistics, error rate, evenness and continuity of coverage, RNA biotype representation, and accuracy of expression profiling. The results were highly reproducible, with relatively little technical variation, and the data generated by the Illumina and Ion Torrent platforms were significantly correlated. The differentially expressed genes identified by RNA-Seq overlapped well with those identified by microarray. However, RNA-Seq detected additional transcripts whose expression levels were either not interrogated or not detected by microarrays.
Fig. 1 shows a brief overview of the miRNA analytical pipeline, divided into 4 stages (Fig. 1). The first step is preparation of the library to select the proper size of RNA, using ≥1 µg according to species. For human samples, the proper range of final library size is 140–160 bp. After library preparation, sequencing is performed. Following NGS of miRNA, filtered and trimmed reads are aligned against the genome using SAM/BAM tools to identify novel and known miRNAs expressed in the sample [34]. Known miRNAs can be identified by comparing and considering major miRNA features, such as sequence conservation among species and structural aspects such as hairpins and minimum folding energy. For novel miRNAs, machine learning algorithms can also be applied. The most commonly used tools for this purpose are miRDeep/miRDeep2 [3536]. These tools are useful for finding both previously known and novel miRNAs, and allow estimation of the accuracy and sensitivity of their performance. Sequencing reads are mapped to the reference genome using bowtie [37]. To obtain information regarding miRNA diversity, expression profiles, and target relationships, several databases are supported with machine learning algorithm to provide the predicted miRNA interaction derived from experimental evidences of miRNA-mRNA interaction [383940]. To take into account several miRNA features and increase prediction efficiency, most databases provide information about significance based on statistical tests. TargetScan is a web-based database that predicts miRNA targets by searching for conserved and nonconserved sites [38]. miRTarBase [39] identifies binding sites for single miRNAs and multiple sites regulated by different miRNAs acting cooperatively. To identify putative miRNA-binding sites even when the targeting miRNA is unknown, RNAhybrid [41] predicts multiple potential miRNA-binding sites in large target RNAs by looking at the most energetically favorable hybridization sites between two separate RNA sequences. DIANA-miRPath can be accessed on the web (http://www.microrna.gr/miRPathv3). This system uses predicted interactions derived from DIANA-microT-CDS [42] and TargetScan6.2, as well as more than 60,000 experimentally supported interactions from DIANA-TarBase-v7.0 [40].
An early miRNA profiling analysis using 27 human bladder specimens comprising 25 urothelial carcinomas and two normal mucosa [21] revealed that ten miRNAs are significantly up-regulated in bladder cancer relative to normal bladder tissue. No down-regulated miRNAs were detected in that study. The miRNAs down-regulated in bladder cancer were first identified using a hybridization-based miRNA array: 14 down-regulated miRNAs were detected, of which four (miR-143, miR-145, miR-125b, and miR-199b) were significantly down-regulated in the same samples [43]. Subsequent studies identified many more miRNAs up- or down-regulated in bladder cancer.
To detection or clarifying early event in tumorigenesis, miRNA expression pattern has been reliable cancer biomarkers and treatment-response predictors for clinical trials since miRNA expression signatures were correlate with tumor classification and were proven useful in determining the primary site of cancers of unknown origin [44]. Pignot [23] demonstrated that expression of three miRNAs (miR-9, miR-182, and miR-200b) is associated with both recurrence-free and overall survival in MIBC. Therefore, these miRNAs can discriminate MIBC tumors in terms of their aggressiveness and potential for recurrence.
To date, 19 research groups have reported 261 miRNAs differentially expressed in bladder cancer, of which 83 were identified in multiple studies. Ultimately, 76 miRNAs (Table 1) were confirmed as consistently deregulated by two or more groups; 7 of the original 83 miRNAs (miR-106b-3p, miR-141-3p, miR-193a-3p, miR-198, miR-200a-3p, miR-31, and miR-370) were excluded due to inconsistent expression patterns between studies (data not shown). The remaining miRNAs were classified as down-regulated (52) or up-regulated (24) in bladder cancer relative to normal bladder tissue (Table 1). NGS data suggest that both the 5p-arm and 3p-arm of mature miRNAs could be generated from a single miRNA hairpin precursor; however, current miRNA databases do not provide comprehensive arm annotation features, potentially resulting in ambiguous and incomplete analyses. There is no bias in the expression pattern of 5p-miRNA vs 3p-miRNA, although miRNAs from -5p and -3p have variable nucleotide compositions. However, -3p expression in diseased samples tends to show a larger degree of dispersion. Therefore, we performed pathway analysis of aberrantly expressed -5p and -3p miRNAs (specifically, 24 down-regulated miRNAs and four up-regulated miRNAs) (Table 2). The functions of these 28 miRNAs were mainly related to axon guidance, cancer-associated proteoglycans, and the ErbB and transforming growth factor-beta signaling pathways (Table 3).
Aberrant miRNA expression in urinary sediment or supernatant reflects the states of urothelial cancer cells, based on the inclusion of miRNAs in various protein complexes or membranous particles such as exosomes or microvesicles [585960]. Therefore, previous reports of the miRNA profiling in urine is a attractive information for cancer biomarkers discovery. Comparison of miRNA profiling between 50 miRNAs derived from urine (Table 4) and 190 miRNAs derived from bladder tissue which included 76 multiple reported miRNAs. A Venn diagram revealed that the multiply reported miRNAs expressed in bladder tissues and the miRNAs detected in urine shared 14 miRNAs in common. Therefore, these 14 miRNAs represent promising biomarkers for diagnostic applications (Fig. 2). Furthermore, the 190 miRNAs from bladder cancer tissue and the 50 miRNAs from urine also shared 14 miRNAs in common, providing further support for the idea that these molecules have potential as novel diagnostic and prognostic biomarkers. Finally, 22 miRNAs required further validation to determine whether they were derived from any cells including urothelial carcinoma that may be shed into the urine.
To summarize, we provide evidence that miRNA profiling data could provide important insights into bladder tumorigenesis, and that miRNAs in urine may originate in urothelial cancer cells. Future studies should focus on the mechanisms of association and explore the potential of miRNAs from cancer tissue and urine as valid biomarkers for the diagnosis, classification, and prognosis of bladder cancer.
Figures and Tables
Fig. 2
Venn diagram of single and multiple reported microRNAs (miRNAs) from bladder cancer tissues, and urine. A 28 miRNAs were shared between bladder cancer tissues and urine which included 14 miRNAs of two or more than groups which comparison of 190 miRNAs from bladder cancer tissues including 76 miRNAs of two or more than groups, and 50 miRNAs from urine. (A) miRNAs from bladder cancer tissues, urine and also multiple reports. (B) miRNAs from bladder cancer tissues and urine. (C) miRNA in urine.

Table 1
Seventy-six miRNAs identified in multiple studies in bladder cancer
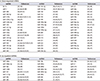
| A. Down-regulated miRNAs | |||||
|---|---|---|---|---|---|
| miRNA | References | miRNA | References | miRNA | References |
| let-7c | [45,46,47,48] | miR-145-3p | [46,47,49,51] | miR-25 | [43,49] |
| miR-1 | [23,36,46,47,48,49] | miR-145-5p | [45,46,47,49,51,52] | miR-26a | [53,56] |
| miR-100 | [46,49,50,51] | miR-150 | [43,50] | miR-26a-5p | [45,52] |
| miR-100-5p | [47,52] | miR-152 | [47,50] | miR-27b-3p | [45,52] |
| miR-125b | [43,46,47,49,50,51,53] | miR-195 | [47,49] | miR-29a | [43,53] |
| miR-125b-2-5p | [46,47] | miR-195 | [43,46,50,51] | miR-29c | [36,53,56] |
| miR-125b-5p | [45,52] | miR-199a-3p | [47,49,51,55] | miR-30a-3p | [50,52] |
| miR-126 | [43,53] | miR-199a-5p | [47,49,50,55] | miR-30a-5p | [45,47,52] |
| miR-127-3p | [47,52] | miR-199b | [23,43] | miR-30e-5p | [52,56] |
| miR-130a | [47,49] | miR-199b-3p | [47,52,55] | miR-320 | [48,50] |
| miR-133a | [23,46,47,48,49,50,54] | miR-204 | [20,23,48] | miR-490-3p | [46,47] |
| miR-133b | [20,23,47,48,49,50,54] | miR-214 | [48,49,51] | miR-490-5p | [46,47] |
| miR-139-5p | [47,48,49,51] | miR-218 | [48,50] | miR-497 | [47,49] |
| miR-140-3p | [48,52] | miR-221 | [43,49] | miR-638 | [45,49] |
| miR-143 | [23,36,43,47,49,51,53] | miR-222 | [49,51] | miR-99a | [49,50] |
| miR-143-3p | [45,46,52] | miR-223 | [47,50] | miR-99a-3p | [46,47] |
| miR-143-5p | [46,47,52] | miR-23b | [43,47] | miR-99a-5p | [46,47] |
| miR-145 | [23,36,43,50,53] | miR-23b-3p | [45,52] | ||
| B. Up-regulated miRNAs | |||||
|---|---|---|---|---|---|
| miRNA | References | miRNA | References | miRNA | References |
| miR-106a | [43,50] | miR-18a | [43,46] | miR-20a | [43,46,51,53] |
| miR-106b-3p | [52,55] | miR-200a | [43,46,49,55] | miR-21 | [20,53] |
| miR-10a | [36,49] | miR-200b | [23,49] | miR-210 | [43,45,49,51] |
| miR-130b | [50,51] | miR-200b-3p | [45,46,52,55] | miR-224 | [36,50,54] |
| miR-141 | [46,49,51,55,57] | miR-200b-5p | [49,52,55] | miR-429 | [46,49,55,57] |
| miR-17-5p | [21,43,46] | miR-200c | [43,46,49,55,57] | miR-93 | [20,43,55] |
| miR-182 | [23,36,46,49,54] | miR-203 | [21,36,54] | miR-96 | [46,50] |
| miR-183 | [36,46,49,50,54] | miR-205 | [21,49,51] | ||
Table 2
Twenty-four down-regulated and 4 up-regulated miRNAs from the set of 76 consistently expressed miRNAs in bladder cancer tissues

Table 3
KEGG pathway analysis of 28 miRNAs

Table 4
Deregulated 50 miRNAs in urine

| miRNA | Reference | miRNA | Reference | miRNA | Reference |
|---|---|---|---|---|---|
| let-7b | [52] | miR-182 | [61] | miR-377 | [62] |
| let-7i | [62] | miR-191 | [52] | miR-451a | [52] |
| miR-100 | [63] | miR-192 | [60] | miR-483 | [62] |
| miR-101 | [62] | miR-200a | [60,64] | miR-505 | [62] |
| miR-10b | [52] | miR-200b | [60] | miR-509 | [62] |
| miR-1224 | [63] | miR-200c | [60] | miR-515 | [62] |
| miR-1255b | [59] | miR-203 | [63] | miR-545 | [62] |
| miR-125a | [52] | miR-205 | [60] | miR-556 | [62] |
| miR-125b | [65] | miR-212 | [63] | miR-589 | [62] |
| miR-126 | [61,65] | miR-214 | [2] | miR-616 | [62] |
| miR-134 | [62] | miR-223 | [52,62] | miR-618 | [59] |
| miR-135b | [63] | miR-24-1 | [63] | miR-873 | [62] |
| miR-145 | [64] | miR-27b | [63] | miR-890 | [62] |
| miR-152 | [61] | miR-302d | [62] | miR-892a | [62] |
| miR-155 | [60] | miR-325 | [62] | miR-923 | [62] |
| miR-15a | [63] | miR-328 | [63] | miR-99a | [52] |
| miR-15b | [63] | miR-335 | [62] |
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012; 62:10–29.
2. Kim SM, Kang HW, Kim WT, Kim YJ, Yun SJ, Lee SC, et al. Cell-free microRNA-214 from urine as a biomarker for nonmuscle-invasive bladder cancer. Korean J Urol. 2013; 54:791–796.
3. Messing E. Markers of detection. Urol Oncol. 2007; 25:344–347.
4. Quan C, Cha EJ, Lee HL, Han KH, Lee KM, Kim WJ. Enhanced expression of peroxiredoxin I and VI correlates with development, recurrence and progression of human bladder cancer. J Urol. 2006; 175:1512–1516.
5. Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Bohle A, Palou-Redorta J, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder. Eur Urol. 2008; 54:303–314.
6. Sylvester RJ. Natural history, recurrence, and progression in superficial bladder cancer. ScientificWorldJournal. 2006; 6:2617–2625.
7. Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009; 374:239–249.
8. Knowles MA. Role of FGFR3 in urothelial cell carcinoma: biomarker and potential therapeutic target. World J Urol. 2007; 25:581–593.
9. Pandith AA, Shah ZA, Siddiqi MA. Oncogenic role of fibroblast growth factor receptor 3 in tumorigenesis of urinary bladder cancer. Urol Oncol. 2013; 31:398–406.
10. Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer. 2005; 5:713–725.
11. Bakkar AA, Wallerand H, Radvanyi F, Lahaye JB, Pissard S, Lecerf L, et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer Res. 2003; 63:8108–8112.
12. Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999; 23:18–20.
13. Wallerand H, Reiter RR, Ravaud A. Molecular targeting in the treatment of either advanced or metastatic bladder cancer or both according to the signalling pathways. Curr Opin Urol. 2008; 18:524–532.
14. Cazier JB, Rao SR, McLean CM, Walker AK, Wright BJ, Jaeger EE, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun. 2014; 5:3756.
15. Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3' untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009; 16:144–150.
16. Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004; 5:396–400.
17. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006; 94:776–780.
18. Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs: the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009; 9:293–302.
19. Bandres E, Agirre X, Ramirez N, Zarate R, Garcia-Foncillas J. MicroRNAs as cancer players: potential clinical and biological effects. DNA Cell Biol. 2007; 26:273–282.
20. Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009; 69:8472–8481.
21. Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007; 25:387–392.
22. Hidaka H, Seki N, Yoshino H, Yamasaki T, Yamada Y, Nohata N, et al. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget. 2012; 3:44–57.
23. Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A, et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. Int J Cancer. 2013; 132:2479–2491.
24. Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013; 10:396–404.
25. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008; 18:1509–1517.
26. Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010; 464:773–777.
27. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008; 5:621–628.
28. Mutz KO, Heilkenbrinker A, Lonne M, Walter JG, Stahl F. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol. 2013; 24:22–30.
29. Nagalakshmi U, Waern K, Snyder M. RNA-Seq: a method for comprehensive transcriptome analysis. Curr Protoc Mol Biol. 2010; Chapter 4:Unit 4.11.1-13.
30. Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011; 8:469–477.
31. Bottomly D, Walter NA, Hunter JE, Darakjian P, Kawane S, Buck KJ, et al. Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS One. 2011; 6:e17820.
32. Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, et al. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:10084–10097.
33. Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014; 9:e78644.
34. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079.
35. Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012; 40:37–52.
36. Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009; 69:2623–2629.
37. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012; 9:357–359.
38. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015; 4:e05005.
39. Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016; 44:D239–D247.
40. Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015; 43:W460–W466.
41. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10:1507–1517.
42. Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013; 41:W169–W173.
43. Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C, et al. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009; 181:1372–1380.
44. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005; 435:834–838.
45. Canturk KM, Ozdemir M, Can C, Oner S, Emre R, Aslan H, et al. Investigation of key miRNAs and target genes in bladder cancer using miRNA profiling and bioinformatic tools. Mol Biol Rep. 2014; 41:8127–8135.
46. Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011; 6:e18286.
47. Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, et al. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLoS One. 2014; 9:e84311.
48. Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011; 104:808–818.
49. Song T, Xia W, Shao N, Zhang X, Wang C, Wu Y, et al. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev. 2010; 11:905–911.
50. Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009; 125:345–352.
51. Ratert N, Meyer HA, Jung M, Lioudmer P, Mollenkopf HJ, Wagner I, et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J Mol Diagn. 2013; 15:695–705.
52. Chen YH, Wang SQ, Wu XL, Shen M, Chen ZG, Chen XG, et al. Characterization of microRNAs expression profiling in one group of Chinese urothelial cell carcinoma identified by Solexa sequencing. Urol Oncol. 2013; 31:219–227.
53. Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009; 69:4851–4860.
54. Zhu J, Jiang Z, Gao F, Hu X, Zhou L, Chen J, et al. A systematic analysis on DNA methylation and the expression of both mRNA and microRNA in bladder cancer. PLoS One. 2011; 6:e28223.
55. Li X, Chen J, Hu X, Huang Y, Li Z, Zhou L, et al. Comparative mRNA and microRNA expression profiling of three genitourinary cancers reveals common hallmarks and cancer-specific molecular events. PLoS One. 2011; 6:e22570.
56. Wang G, Zhang H, He H, Tong W, Wang B, Liao G, et al. Upregulation of microRNA in bladder tumor tissue is not common. Int Urol Nephrol. 2010; 42:95–102.
57. Jia AY, Castillo-Martin M, Bonal DM, Sanchez-Carbayo M, Silva JM, Cordon-Cardo C. MicroRNA-126 inhibits invasion in bladder cancer via regulation of ADAM9. Br J Cancer. 2014; 110:2945–2954.
58. Cheng Y, Deng X, Yang X, Li P, Zhang X, Li P, et al. Urine microRNAs as biomarkers for bladder cancer: a diagnostic meta-analysis. Onco Targets Ther. 2015; 8:2089–2096.
59. Tolle A, Jung M, Rabenhorst S, Kilic E, Jung K, Weikert S. Identification of microRNAs in blood and urine as tumour markers for the detection of urinary bladder cancer. Oncol Rep. 2013; 30:1949–1956.
60. Wang G, Chan ES, Kwan BC, Li PK, Yip SK, Szeto CC, et al. Expression of microRNAs in the urine of patients with bladder cancer. Clin Genitourin Cancer. 2012; 10:106–113.
61. Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010; 28:655–661.
62. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010; 56:1733–1741.
63. Miah S, Dudziec E, Drayton RM, Zlotta AR, Morgan SL, Rosario DJ, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012; 107:123–128.
64. Yun SJ, Jeong P, Kim WT, Kim TH, Lee YS, Song PH, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol. 2012; 41:1871–1878.
65. Snowdon J, Boag S, Feilotter H, Izard J, Siemens DR. A pilot study of urinary microRNA as a biomarker for urothelial cancer. Can Urol Assoc J. 2013; 7:28–32.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download