Abstract
Background
Accurate measurement of the concentration of hepatitis B virus (HBV) DNA in clinical samples is important for the appropriate treatment of patients and evaluation of their therapeutic responses. In addition, the concentration of HBV DNA in the serum of patients with chronic HBV infection has a very broad range. Real-time PCR is very sensitive and useful to detect HBV DNA in a wide range of concentrations. We designed new primers and probes for real-time PCR to detect HBV DNA.
Methods
Primers and probes specific for HBV were designed. EUROHEP HBV DNA standards (NIBSC, Hertfordshire, UK) with the HBV DNA concentration of 7.0×104 copies/mL was used to determine the calibration curve and efficacy for the real-time PCR assay. Sensitivity, dynamic range, and precision were evaluated. The correlation between the real-time PCR and Cobas Amplicor HBV Monitor™ assay in the measurement of serum HBV DNA concentrations in 52 patients with chronic HBV infection was evaluated.
Results
The sensitivity of the assay was approximately 6.08×102 copies/mL for HBV, and the quantitation was accurate and reproducible over a wide dynamic range from 6.1×02 to 6.5×109 copies/mL without any dilution of specimens. The assay showed low coefficients of variation of repeatability (3.7–24.9%) and reproducibility (7.8–24.7%). The results were found to correlate well with those obtained by Cobas Amplicor HBV Monitor™ kit.
References
1. Hoofnagle JH, Shafritz DA, Popper H. Chronic type B hepatitis and the “healthy” HBsAg carrier state. Hepatology. 1987; 7:758–63.

2. Kaneko S, Miller RH, Di Bisceglie AM, Feinstone SM, Hoofnagle JH, Purcell RH. Detection of hepatitis B virus DNA in serum by polymerase chain reaction. Application for clinical diagnosis. Gastroenterology. 1990; 99:799–804.
3. Hadziyannis SJ, Vassilopoulos D. Hepatitis B e antigen-negative chronic hepatitis B. Hepatology. 2001; 34:617–24.

5. Saldanha J, Gerlich W, Lelie N, Dawson P, Heermann K, Heath A, et al. An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sang. 2001; 80:63–71.

6. Aspinall S, Steele AD, Peenze I, Mphahlele MJ. Detection and quantitation of hepatitis B virus DNA: comparison of two commercial hybridization assays with polymerase chain reaction. J Viral Hepat. 1995; 2:107–11.

7. Barlet V, Cohard M, Thelu MA, Chaix MJ, Baccard C, Zarski JP, et al. Quantitative detection of hepatitis B virus DNA in serum using chemiluminescence: comparison with radioactive solution hybridization assay. J Virol Methods. 1994; 49:141–51.

8. Krajden M, Minor J, Cork L, Comanor L. Multi-measurement method comparison of three commercial hepatitis B virus DNA quantification assays. J Viral Hepat. 1998; 5:415–22.

9. Chan HL, Leung NW, Lau TC, Wong ML, Sung JJ. Comparison of three different sensitive assays for hepatitis B virus DNA in monitoring of responses to antiviral therapy. J Clin Microbiol. 2000; 38:3205–8.

10. Hendricks DA, Stowe BJ, Hoo BS, Kolberg J, Irvine BD, Neuwald PD, et al. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am J Clin Pathol. 1995; 104:537–46.

11. Ho SK, Chan TM, Cheng IK, Lai KN. Comparison of the second-generation digene hybrid capture assay with the branched-DNA assay for measurement of hepatitis B virus DNA in serum. J Clin Microbiol. 1999; 37:2461–5.

12. Niesters HG, Krajden M, Cork L, de Medina M, Hill M, Fries E, et al. A multicenter study evaluation of the digene hybrid capture II signal amplification technique for detection of hepatitis B virus DNA in serum samples and testing of EUROHEP standards. J Clin Microbiol. 2000; 38:2150–5.

13. Loeb KR, Jerome KR, Goddard J, Huang M, Cent A, Corey L. High-throughput quantitative analysis of hepatitis B virus DNA in serum using the TaqMan fluorogenic detection system. Hepatology. 2000; 32:626–9.

14. Noborg U, Gusdal A, Horal P, Lindh M. Levels of viraemia in subjects with serological markers of past or chronic hepatitis B virus infection. Scand J Infect Dis. 2000; 32:249–52.
15. Michalak TI, Pasquinelli C, Guilhot S, Chisari FV. Hepatitis B virus persistence after recovery from acute viral hepatitis. J Clin Invest. 1994; 93:230–9.

16. Lok AS, Heathcote EJ, Hoofnagle JH. Management of hepatitis B: 2000-summary of a workshop. Gastroenterology. 2001; 120:1828–53.

17. Martinot-Peignoux M, Boyer N, Colombat M, Akremi R, Pham BN, Ollivier S, et al. Serum hepatitis B virus DNA levels and liver histology in inactive HBsAg carriers. J Hepatol. 2002; 36:543–6.

18. Heo J, Baik TH, Kim HH, Kim GH, Kang DH, Song GA, et al. Serum hepatitis B virus (HBV) DNA levels at different stages of clinical course in patients with chronic HBV infection in an endemic area. J Korean Med Sci. 2003; 18:686–90.
19. Oh SH, Kim HH, Heo J, Cho M, Chang CH, Lee EY, et al. Serum hepatitis B virus DNA quantitive analysis using polymerase chain reaction in patients with chronic hepatitis B virus infection. Korean J Lab Med. 2003; 23:39–44.
20. Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, et al. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol. 1999; 37:2899–903.

21. Pas SD, Fries E, De Man RA, Osterhaus AD, Niesters HG. Development of a quantitative real-time detection assay for hepatitis B virus DNA and comparison with two commercial assays. J Clin Microbiol. 2000; 38:2897–901.

22. Weinberger KM, Wiedenmann E, Bohm S, Jilg W. Sensitive and accurate quantitation of hepatitis B virus DNA using a kinetic fluorescence detection system (TaqMan PCR). J Virol Methods. 2000; 85:75–82.

23. Zanella I, Rossini A, Domenighini D, Albertini A, Cariani E. Quantitative analysis of hepatitis B virus DNA by real-lime amplification. Eur J Clin Microbiol Infect Dis. 2002; 21:22–6.
Fig. 2.
Sigmoid fluorescence curves obtained with serial dilutions of standard plasmid. A, distilled water; B, 5.0×109 copies/mL; C, 5.0×107 copies/mL; D, 5.0×105 copies/mL; and E, 5.0×103 copies/mL.
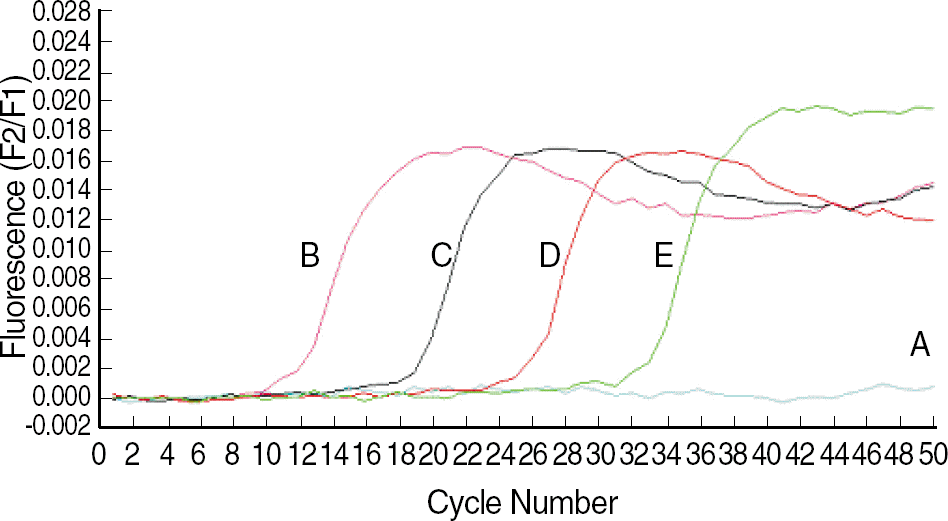
Fig. 3.
The dilutional linearity of HBV DNA quantitation using realtime PCR. The standard curve was obtained with serial dilutions of standard plasmid (♦, mean of 5 experiments).
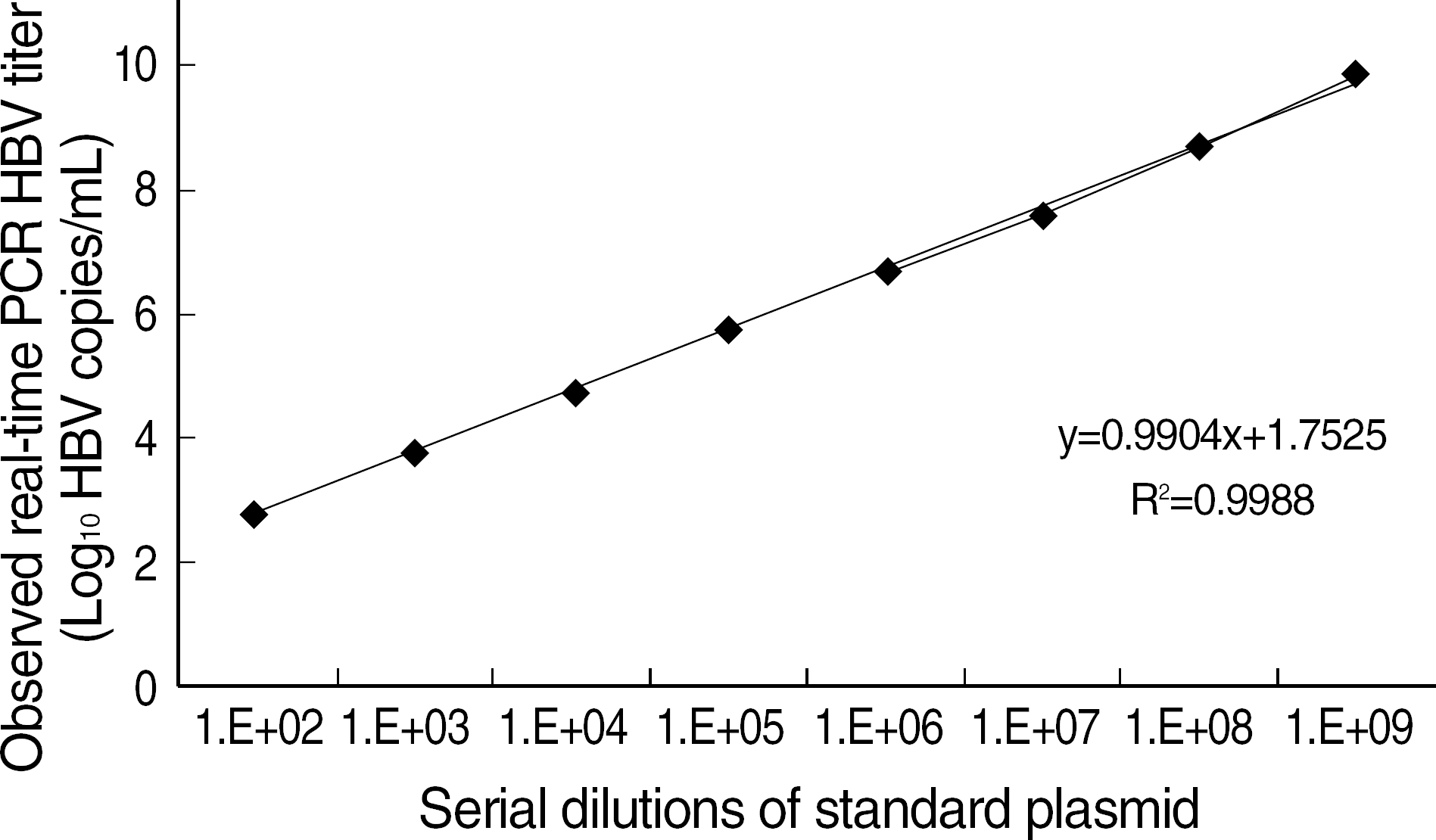
Fig. 4.
Correlation between real-time PCR HBV DNA quantitation and the Cobas Amplicor HBV Monitor™, test (P<0.001). Regression analysis of 52 sera of chronic hepatitis B patients analysed by both test methods. The real-time PCR HBV DNA quantitation test was duplicated. All specimens had titers above 1,000 copies/mL. Samples above 2.0×105 copies/mL in the the Cobas Amplicor HBV Monitor™ test were diluted.
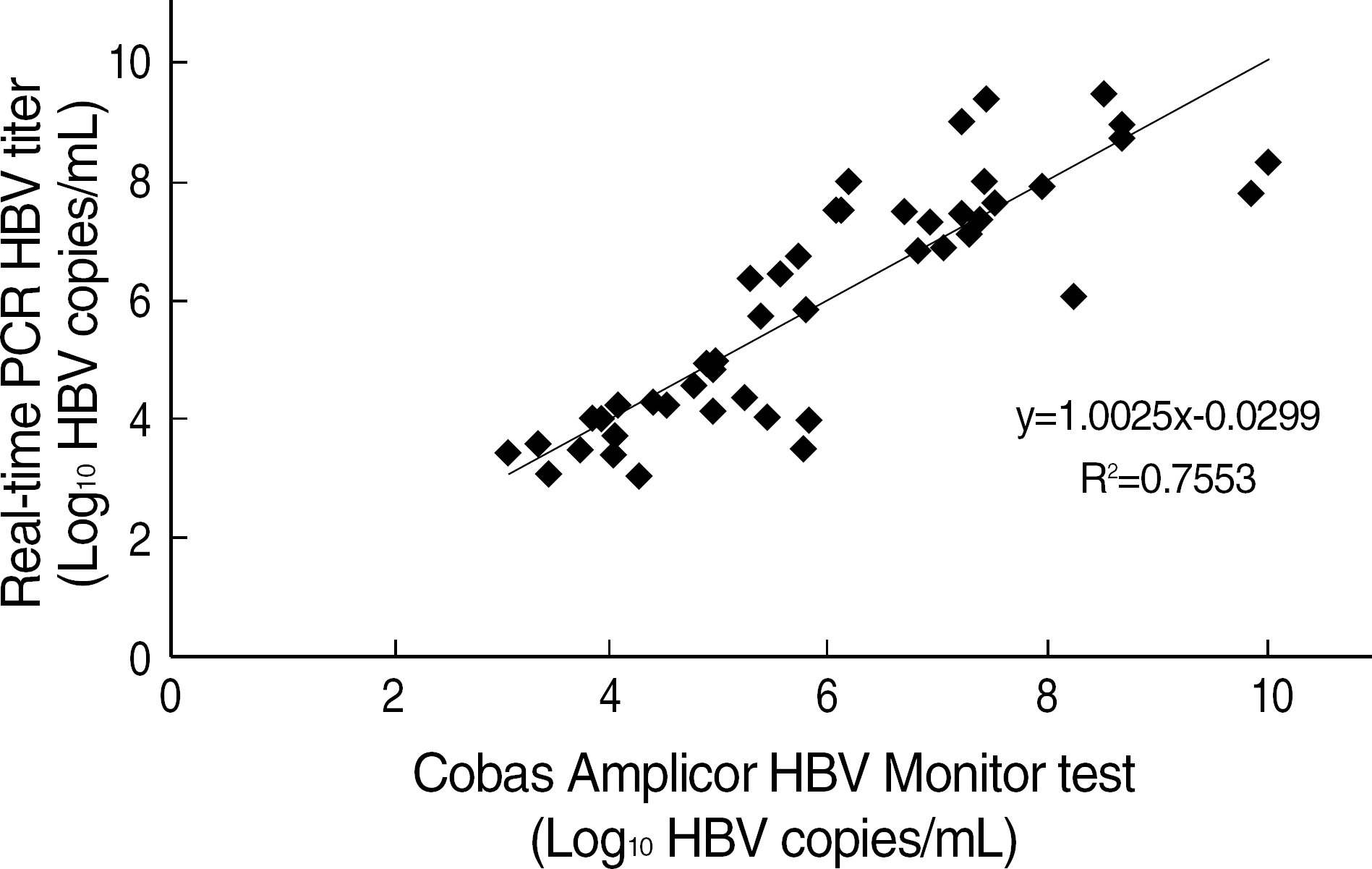
Table 1.
Clinical and laboratory features of patients
Table 2.
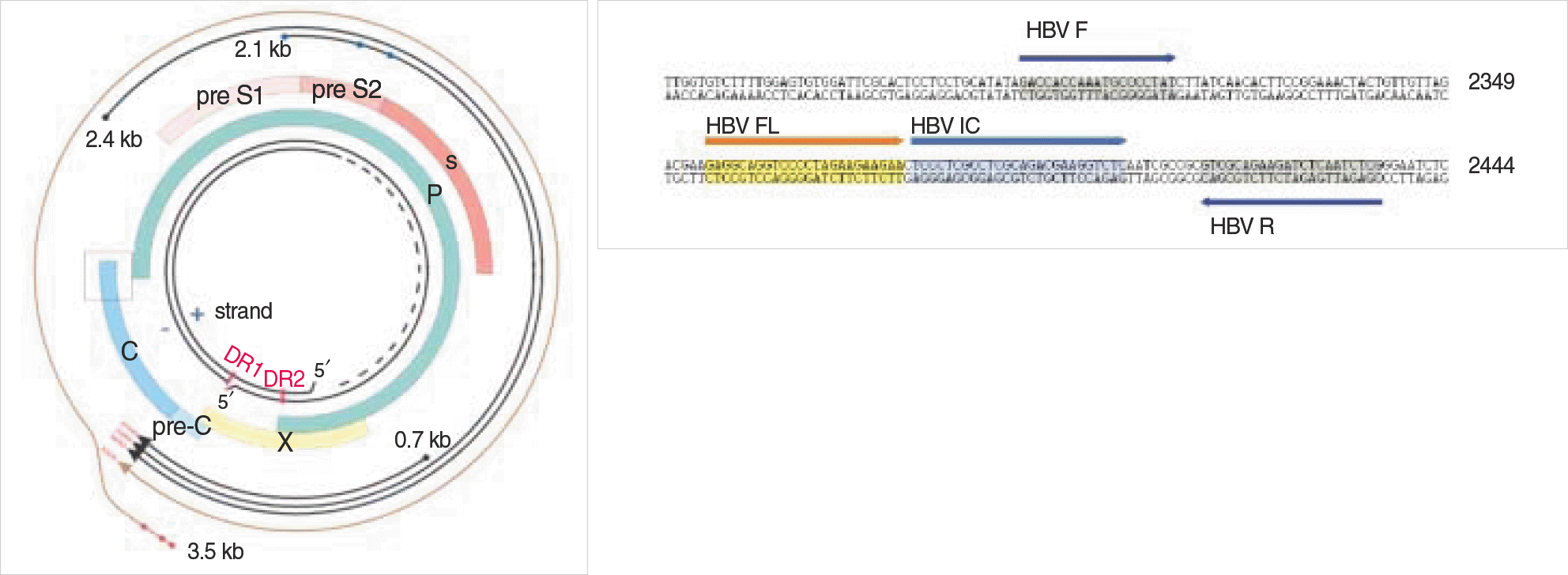
Primers of PCR amplication for standard plasmid
| Primer |
| P1-Eco, forward, 5′-GAGAGAATTCTTTTTCACCTCTGCCTAATCA-3′ |
| P2-Pst, reverse, 5′-GAGACTGCAGAAAAAGTTGCATGGTGCTGG-3′ |
Table 3.
Primers and probes of real-time PCR for HBV DNA quantitation
Table 4.
Precision of real-time PCR on serial dilution of standard plasmid (5.0×108 copies/mL)




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download