Abstract
Here in this study, we investigated the lifespan-extending effect and underlying mechanism of methanolic extract of Moringa olelifa leaves (MML) using Caenorhabditis elegans (C. elegans) model system. To define the longevity properties of MML we conducted lifespan assay and MML showed significant increase in lifespan under normal culture condition. In addition, MML elevated stress tolerance of C. elegans to endure against thermal, oxidative and osmotic stress conditions. Our data also revealed that increased activities of antioxidant enzymes and expressions of stress resistance proteins were attributed to MML-mediated enhanced stress resistance. We further investigated the involvement of MML on the aging-related factors such as growth, food intake, fertility, and motility. Interestingly, MML significantly reduced growth and egg-laying, suggesting these factors were closely linked with MML-mediated longevity. We also observed the movement of aged worms to estimate the effects of MML on the health span. Herein, MML efficiently elevated motility of aged worms, indicating MML may affect health span as well as lifespan. Our genetic analysis using knockout mutants showed that lifespan-extension activity of MML was interconnected with several genes such as skn-1, sir-2.1, daf-2, age-1 and daf-16. Based on these results, we could conclude that MML prolongs the lifespan of worms via activation of SKN-1 and SIR-2.1 and inhibition of insulin/IGF pathway, followed by DAF-16 activation.
REFERENCES
(1). Castillo-Quan J. I., Kinghorn K. J., Bjedov I.Adv. Genet. 2015; 90:1–101.
(2). Stohs S. J., Hartman M. J.Phytother. Res. 2015; 29:796–804.
(3). Anwar F., Latif S., Ashraf M., Gilani A. H.Phytother. Res. 2007; 21:17–25.
(4). Kumar Gupta S., Kumar B., Srinivasan B. P., Nag T. C., Srivastava S., Saxena R., Aggarwal A. J.Ocul. Pharmacol. Ther. 2013; 29:419–426.
(5). Ndhlala A. R., Mulaudzi R., Ncube B., Abdelgadir H. A., du Plooy C. P., Van Staden J.Molecules. 2014; 19:10480–10494.
(6). Waterman C., Cheng D. M., Rojas-Silva P., Poulev A., Dreifus J., Lila M. A., Raskin I.Phytochemistry. 2014; 103:114–122.
(7). Sashidhara K. V., Rosaiah J. N., Tyagi E., Shukla R., Raghubir R., Rajendran S. M.Eur. J. Med. Chem. 2009; 44:432–436.
(8). Bass T. M., Weinkove D., Houthoofd K., Gems D., Partridge L.Mech. Ageing Dev. 2007; 128:546–552.
(9). Abbas S., Wink M.Planta Med. 2009; 75:216–221.
(10). Saini R. K., Shetty N. P., Prakash M., Giridhar P. J.Food Sci. Technol. 2014; 51:2176–2182.
(11). Sadowska-Bartosz I., Bartosz G.Biomed Res. Int. 2014; 2014:404680.
(12). Brenner S.Genetics. 1974; 77:71–94.
(13). Lithgow G. J., White T. M., Melov S., Johnson T. E.Proc. Natl. Acad. Sci. U. S. A. 1995; 92:7540–7544.
(14). Lee E. Y., Shim Y. H., Chitwood D. J., Hwang S. B., Lee J., Paik Y. K.Biochem. Biophys. Res. Commun. 2005; 328:929–936.
(15). Mekheimer R. A., Sayed A. A., Ahmed E. A. J.Med. Chem. 2012; 55:4169–4177.
(16). Horikawa M., Sakamoto K.Biochem. Biophys. Res. Commun. 2009; 390:1402–1407.
(17). Aebi H.Methods Enzymol. 1984; 105:121–126.
(18). Kenyon C. J.Nature. 2010; 464:504–512.
(19). Krishnamurthy P. T., Vardarajalu A., Wadhwani A., Patel V.Indian J. Exp. Biol. 2015; 53:98–103.
(20). Swindell W. R.Aging (Albany NY). 2009; 1:573–577.
(21). Solomon A., Bandhakavi S., Jabbar S., Shah R., Beitel G. J., Morimoto R. I.Genetics. 2004; 167:161–170.
(22). Liu J., Zhang B., Lei H., Feng Z., Liu J., Hsu A. L., Xu X. Z.Cell Metab. 2013; 18:392–402.
(23). Partridge L., Gems D., Withers D. J.Cell. 2005; 120:461–472. 2005.
(24). Bordone L., Guarente L.Nat. Rev. Mol. Cell Biol. 2005; 6:298–305.
(26). Bokov A., Chaudhuri A., Richardson A.Mech. Ageing Dev. 2004; 125:811–826.
(27). Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., Ahringer J., Li H., Kenyon C.Nature. 2003; 424:277–283.
(28). Schaffitzel E., Hertweck M.Exp. Gerontol. 2006; 41:557–563.
(29). Tissenbaum H. A., Guarente L.Nature. 2001; 410:227–230.
(30). Berdichevsky A., Viswanathan M., Horvitz H. R., Guarente L.Cell. 2006; 125:1165–1177.
(31). Mouchiroud L., Houtkooper R. H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y. S., Viswanathan M., Schoonjans K., Guarente L., Auwerx J.Cell. 2013; 154:430–441.
(32). Adachi H., Ishii N. J.Gerontol. A Biol. Sci. Med. Sci. 2000; 55:B280–B285.
(33). Oh S. W., Mukhopadhyay A., Svrzikapa N., Jiang F., Davis R. J., Tissenbaum H. A.Proc. Natl. Acad. Sci. U. S. A. 2005; 102:4494–4499.
(34). Antebi A.Nature. 2007; 447:536–537.
Fig. 1.
Effects of MML on the lifespan of wild-type N2. The number of worms used per each lifespan assay experiment was 40-50 and three independently experiments were repeated (N=3).
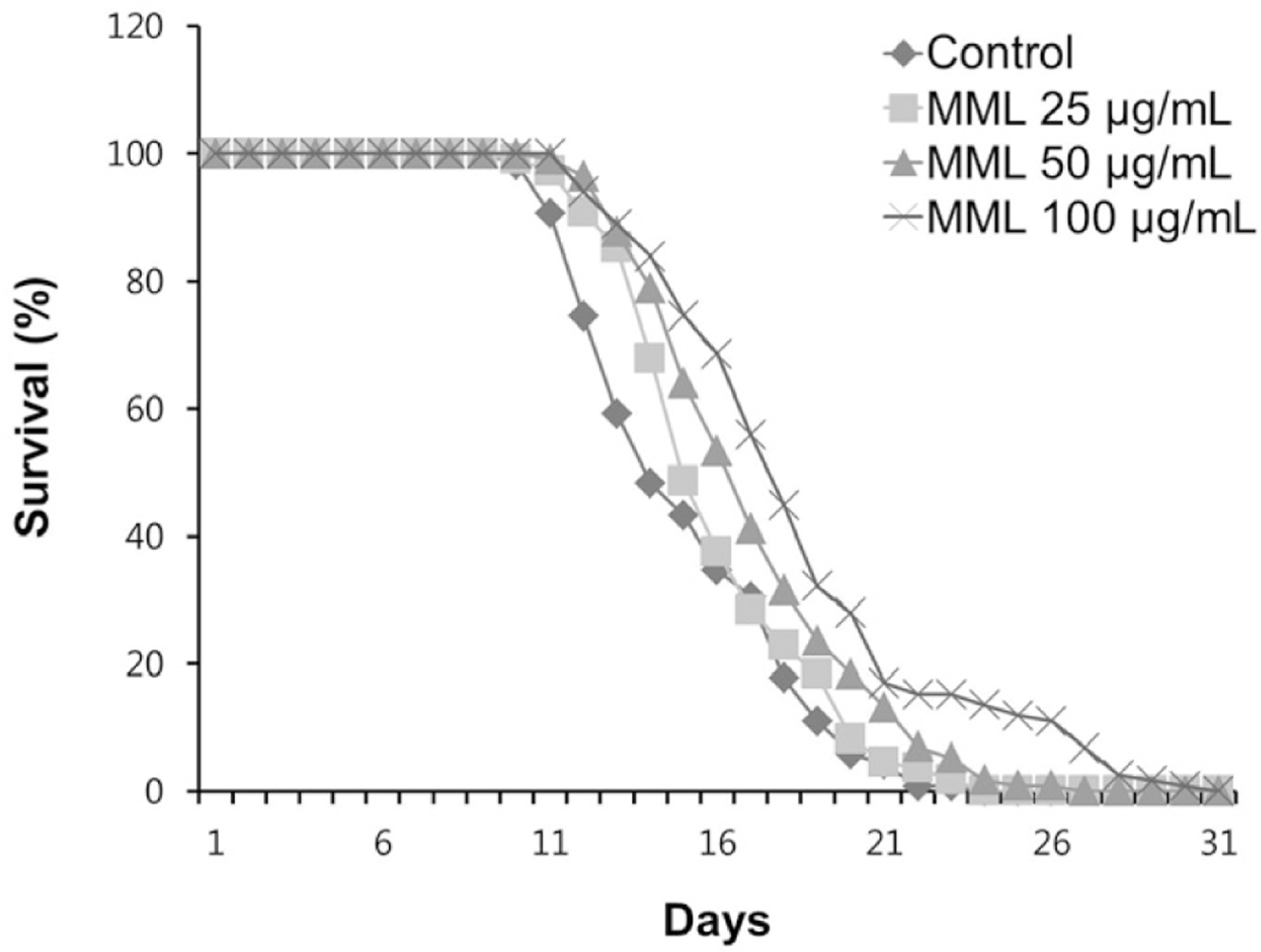
Fig. 2.
Effects of MML on the stress resistance of wild-type N2. (A) To assess thermotolerance, worms were incubated at 36o C and then their viability was scored. (B) For the oxidative stress assay, worms were transferred to NGM agar plate containing 60 mM paraquat, and then their viability was scored. (C) Resistance to osmotic stress was measured by placing 500 mM NaCl and survival rate was calculated.(D) Survival rate of osr-1 (rm1) mutants was recorded under the same osmotic stress condition.
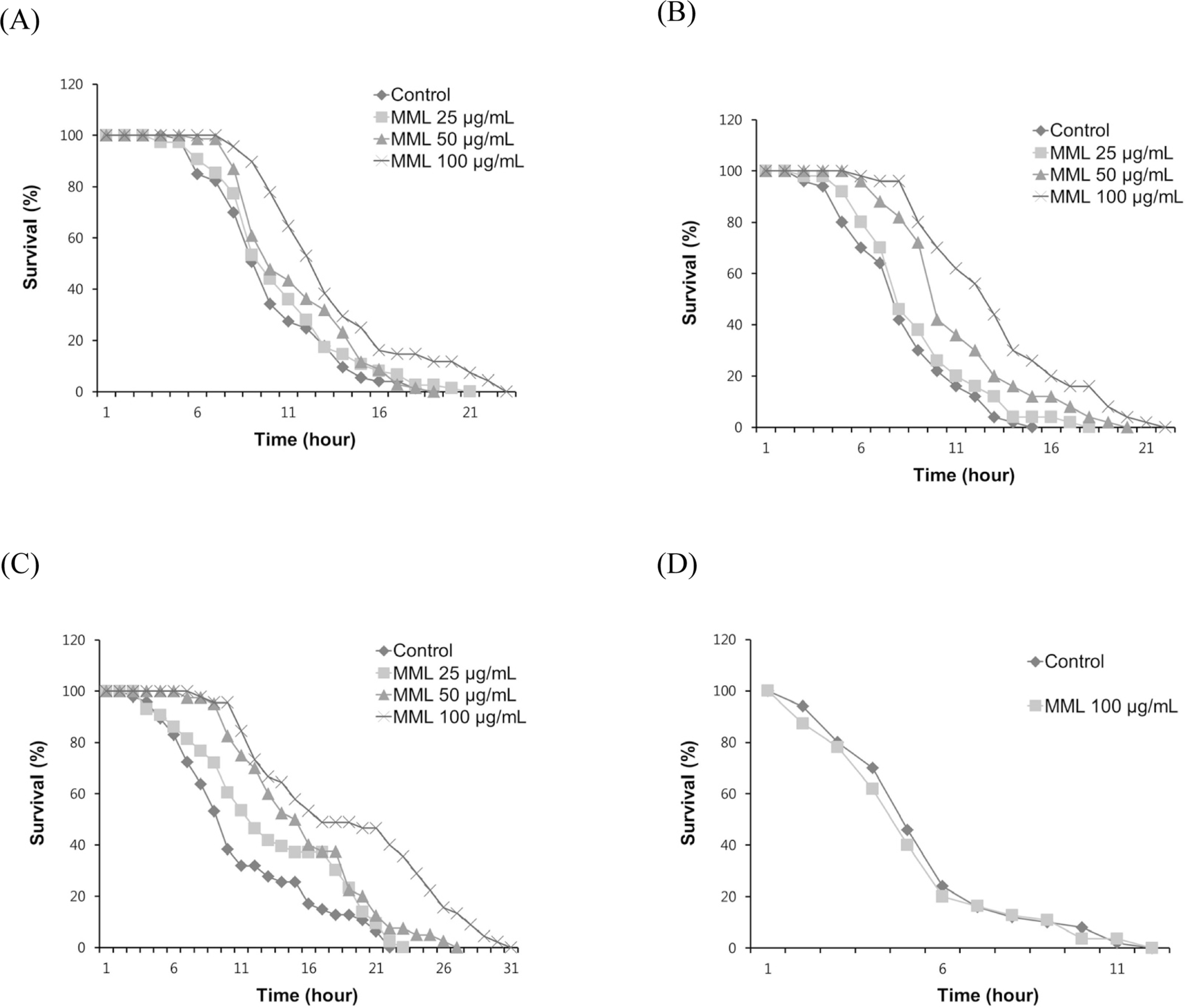
Fig. 3.
Effects of MML on the antioxidant enzyme activity and intracellular ROS levels of wild-type N2. (A-B) Enzyme activity was expressed as a percentage of of the scavenged amount per control. (C) Intracellular ROS accumulation was quantified spec-trophotometrically at excitation 485 nm and emission 535 nm. Plates were read every 30 min for 2 h. All data was expressed as the mean ± S.E.M. of three independent experiments (N=3). Differences compared to the control were considered significant at∗P < 0.05,∗∗P < 0.01 and∗∗∗P < 0.001 by one-way ANOVA.
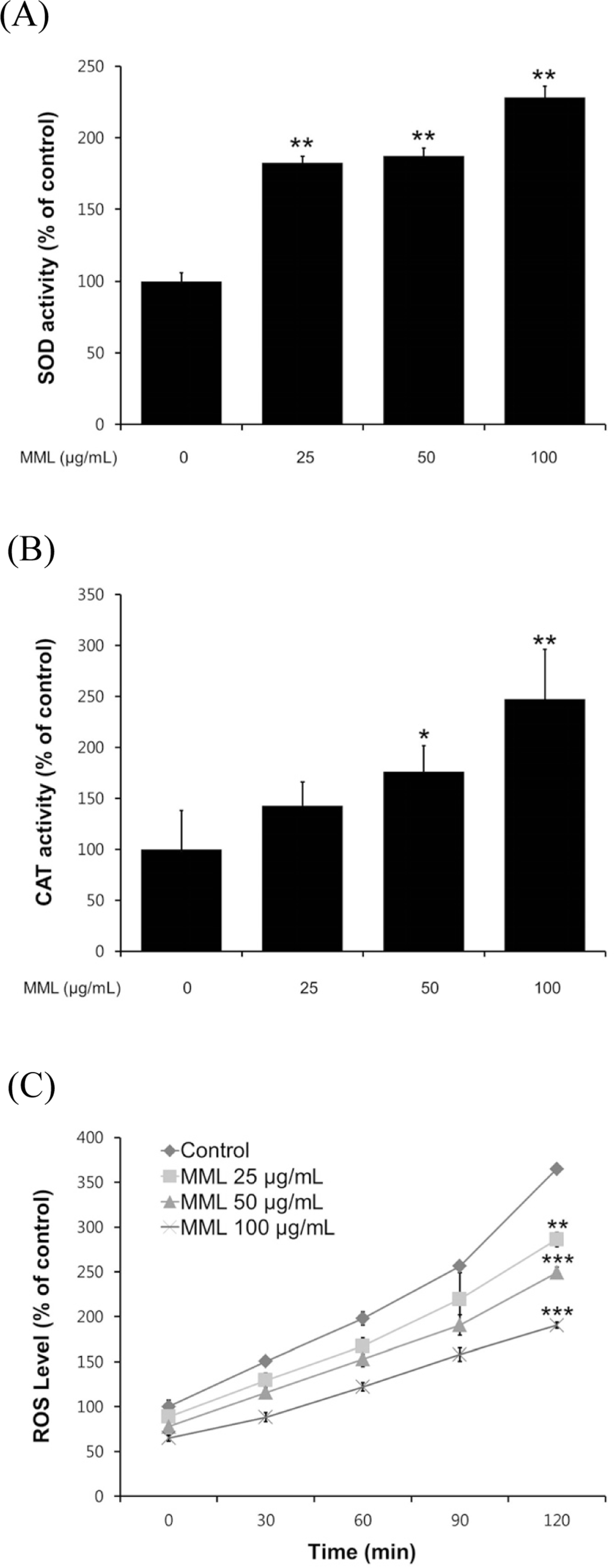
Fig. 4.
Effects of MML on the various aging-related factors of wild-type N2. (A) For the grown alteration assay, photographs were taken on 4th days of adulthood worms. (B) On the 4th days of adulthood, the pharyngeal pumping rates were counted under a dissecting microscope for 1 min. (C) To check the body movement, the analysis was taken on 8th days of adulthood worms. Both assessments were analyzed by the Nikon software (Nikon, Japan). (D) In order to fertility assay, daily and total laid eggs were counted. The progeny was counted at the L2 or L3 stage. Data are expressed as the mean ± S.E.M. of three independent experiments (N=3). Differences compared to the control were considered significant at∗P < 0.05 and∗∗P < 0.01 by one-way ANOVA.
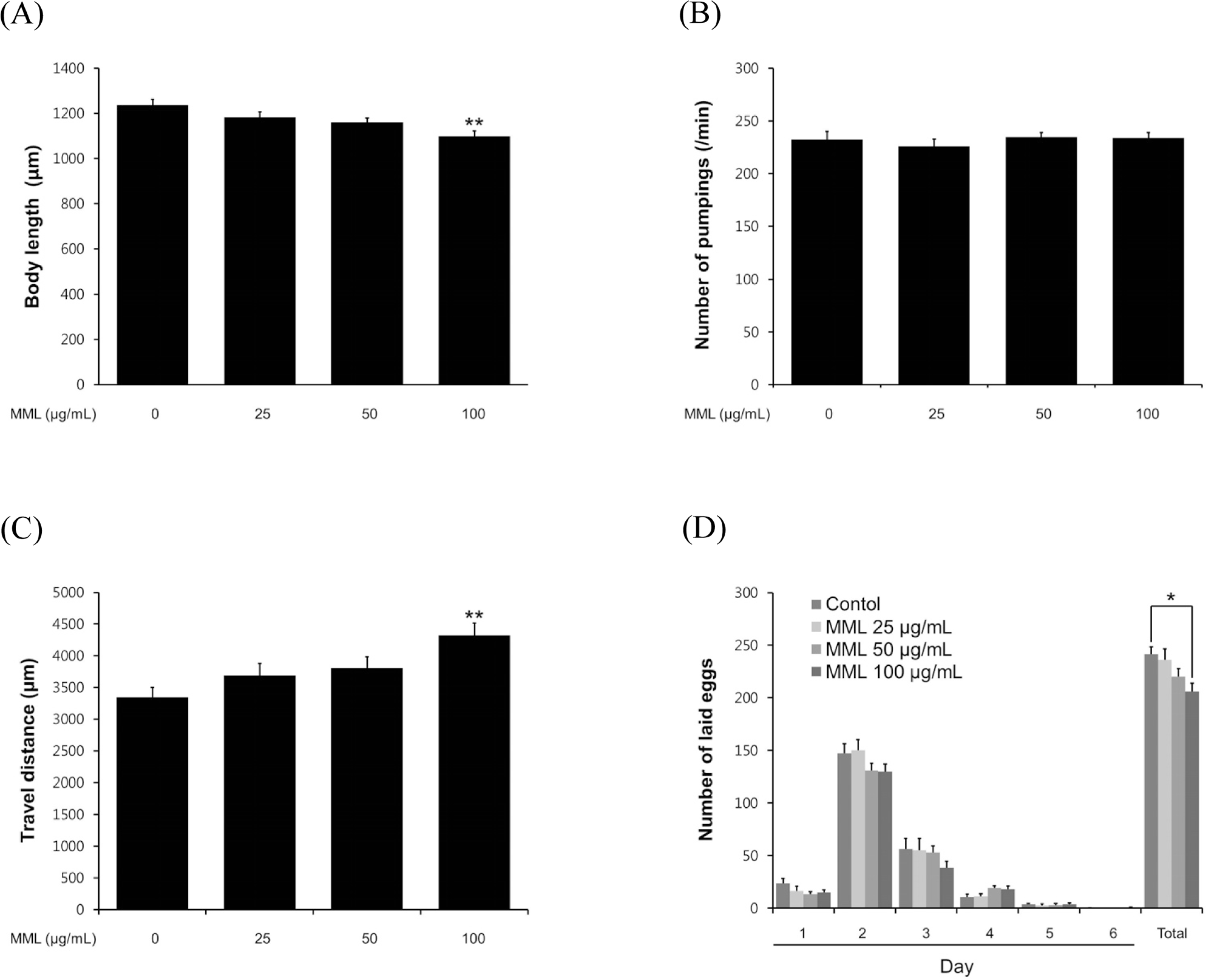
Fig. 5.
Effects of MML on expression of HSP-16.1 and SOD-3 in C. elegans (CL2070 and CF1553 Respectively). (A) Image of HSP-16.1::GFP expression in control and MML-treated worms.(B) Image of SOD-3::GFP expression in control and MML-treated worms. The GFP intensity was quantified using Image J software by determining average pixel intensity. Data are expressed as the mean ± S.E.M. of three independent experiments (N=3). Differences compared to the control were considered significant at∗∗∗P < 0.001 by one-way ANOVA.
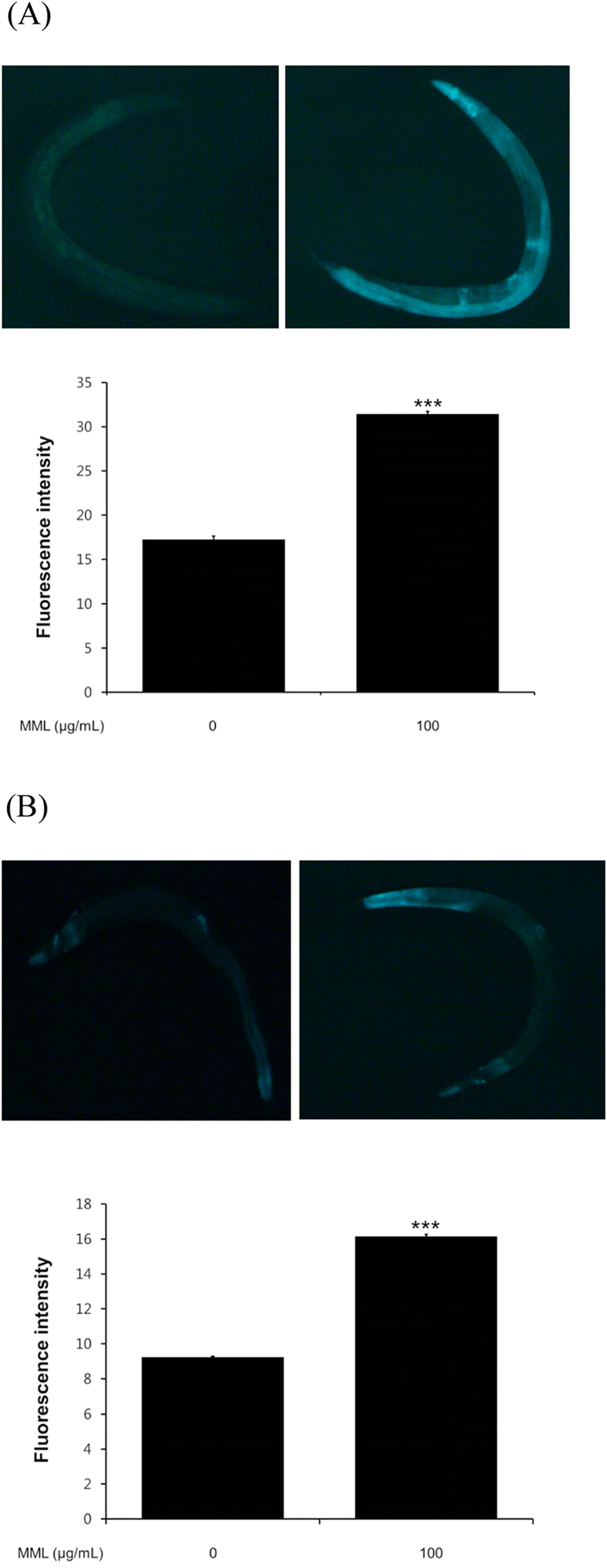
Table 1.
Effects of MML on the lifespan of wild-type N2 and various mutants.
| Genotype | Mean lifespana | Maximum lifespan | Change in mean lifespanc (%) | Log-lank Testd | ||
|---|---|---|---|---|---|---|
| Untreated | Treatedb | Untreated | Treatedb | |||
| Wild-type | 14.2 ± 0.3 | 17.7 ± 0.4 | 23 | 30 | 24.6% | ∗∗∗ p < 0.001 |
| skn-1 (zu67) | 11.6 ± 0.4 | 11.0 ± 0.5 | 21 | 19 | -5.2% | p = 0.965 |
| daf-16 (mgDf50) | 12.6 ± 0.4 | 12.1 ± 0.4 | 21 | 19 | -4.0% | p = 0.758 |
| daf-2 (el368) | 17.1 ± 0.5 | 16.9 ± 0.7 | 29 | 27 | -1.2% | p = 0.169 |
| age-1 (hx546) | 17.4 ± 0.5 | 17.4 ± 0.5 | 27 | 28 | -0.5% | p = 0.252 |
| sir-2.1 (ok434) | 12.1 ± 0.4 | 12.4 ± 0.4 | 19 | 20 | 2.5% | p = 0.240 |
| mek-1 (ks54) | 10.9 ± 0.3 | 12.0 ± 0.4 | 18 | 22 | 10.1% | p = 0.006 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download