This article has been
cited by other articles in ScienceCentral.
Abstract
Noncompartmental analysis (NCA) is a primary analytical approach for pharmacokinetic studies, and its parameters act as decision criteria in bioequivalent studies. Currently, NCA is usually carried out by commercial softwares such as WinNonlin®. In this article, we introduce our newly-developed two R packages, NonCompart (NonCompartmental analysis for pharmacokinetic data) and ncar (NonCompartmental Analysis for pharmacokinetic Report), which can perform NCA and produce complete NCA reports in both pdf and rtf formats. These packages are compatible with CDISC (Clinical Data Interchange Standards Consortium) standard as well. We demonstrate how the results of WinNonlin® are reproduced and how NCA reports can be obtained. With these R packages, we aimed to help researchers carry out NCA and utilize the output for early stages of drug development process. These R packages are freely available for download from the CRAN repository.
Keywords: NonCompart, ncar, R, noncompartmental analysis (NCA)
Introduction
The aim of pharmacokinetic (PK) studies is to examine the kinetics of a drug with regard to absorption, distribution, metabolism and elimination in the body. PK data analysis consists of noncompartmental analysis (NCA) and nonlinear regression analysis.[
12] NCA uses the trapezoidal rule for measurement of area under the concentration-time curve (AUC), and requires fewer assumptions than model-based analysis.[
2] NCA allows for estimation of various PK parameters such as AUC, peak observed drug concentration (C
max), time of peak concentration (T
max), and elimination half-life. Particularly, AUC and C
max are often accepted as the criteria for approval of bioequivalent drugs.
R, a widely-used computer language, is a suite of libraries of statistical and mathematical computations.[
3] Despite its relatively small base system compared with other commercial softwares for NCA such as WinNonlin®[
4] and Kinetica,[
5] R has robust functions for scientific computation and numerous add-in packages for use in various fields.[
6] Therefore, many efforts are being made to replace commercial softwares with R packages.
In this article, we introduce two newly-developed R packages,
NonCompart and
ncar, that are compatible with SDTM (Study Data Tabulation Model)-formatted dataset of CDISC (Clinical Data Interchange Standards Consortium), which is the standard of documentation submitted to regulatory authorities,[
7] while providing a practical method for producing complete NCA reports.
Methods
R packages (NonCompart and ncar)
Two R packages (NonCompart and ncar) for NCA were developed in the open-source R programming language in order to allow free public use. R packages can be installed and loaded using the following scripts.
install.packages(c("NonCompart","ncar"))
Software
WinNonlin® (Pharsight, Mountain View, CA, USA)[
4] in Microsoft-Windows 7 (64 bit) was used for the computation. R 3.4.2 in Microsoft-Windows 7 (64 bit) was used for the comparison of calculated values.
Dataset
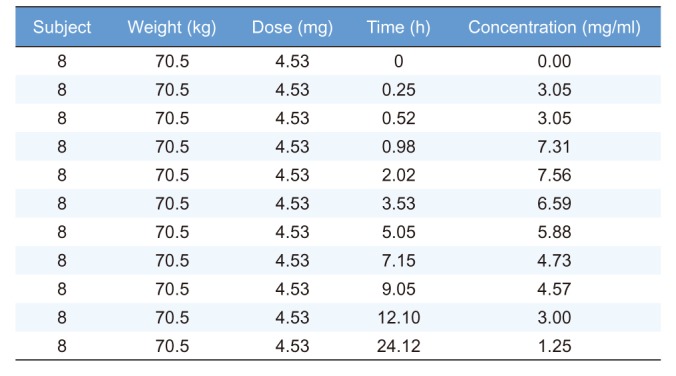
To compare the outputs generated by R packages and WinNonlin®, we used Theoph dataset obtained from the R software. The Theoph dataset has 132 observations from 12 subjects. A portion of the Theoph dataset (subject ID = 8) is shown in
Table 2.
Results
NonCompart package: performance of NCA
This package conducts NCA as similarly as possible to the most widely used commercial PK analysis software. The
NonCompart package has two main functions, tblNCA and sNCA, for use in multiple subjects and one subject, respectively.
Figure 1 shows an example of output by tblNCA. The input data for tblNCA( ) should be in a long format as exemplified by the Theoph dataset.
It is possible to input several keys such as subject demographics and information regarding dose, period, or sequence; the result of tblNCA( ) will print the key columns and the carried keys can be further used for additional statistical analysis (i.e. descriptive statistics, bioequivalence test, t-test, or ANOVA). The adm argument can be 'Extravascular', 'Bolus', or 'Infusion' and the down argument can be either 'Linear' or 'Log'. The greatest advantage of this package is that the outputs produced by this package are compatible with those of pharmacokinetic parameter (PP) TESTCD of CDISC SDTM.
tblNCA(Theoph, key = "Subject", colTime ="Time", colConc = "conc", dose = 320, adm = "Extravascular", dur = 0, doseUnit = "mg", timeUnit = "h", concUnit = "mg/L", down = "Linear")
IntAUC( ) function calculates interval (partial) AUC (from t1 and t2) with the given series of time and concentration. The interval AUC (0.5–11 hour) of the subject 8 can be calculated using the Theoph dataset with the following R script.
Time = Theoph[Theoph$Subject == 8, "Time"]
Concentration = Theoph[Theoph$Subject == 8, "conc"]
Res =sNCA(Time, Concentration, dose = 320, concUnit = "mg/L")
IntAUC(Time, Concentration, t1 = 0.5, t2 = 11, Res)
ncar package: generation of NCA reports
This package generates complete NCA reports including plots with both linear and logarithmic scale. Its two main functions are pdfNCA and rtfNCA, which produce pdf file format and rtf file format, respectively. The generated reports are similar to those generated from commercial softwares, but like
NonCompart, this package has the advantage of using PPTESTCD of CDISC SDTM.
ncar produces NCA reports through
NonCompart and converts them into Microsoft Word format when using rtfNCA( ), which is convenient for editing. Reports generated by pdfNCA( )function show individual plots with trend lines that joins the dots used for calculating terminal slopes.
Figure 2 shows an example of an NCA report in pdf format and an individual concentration-time plot.
pdfNCA(fileName = "pdfNCA-Theoph.pdf", Theoph, colSubj = "Subject", colTime = "Time", colConc = "conc", dose = 320, doseUnit = "mg", timeUnit = "h", concU nit = "mg/L", down = "Linear")
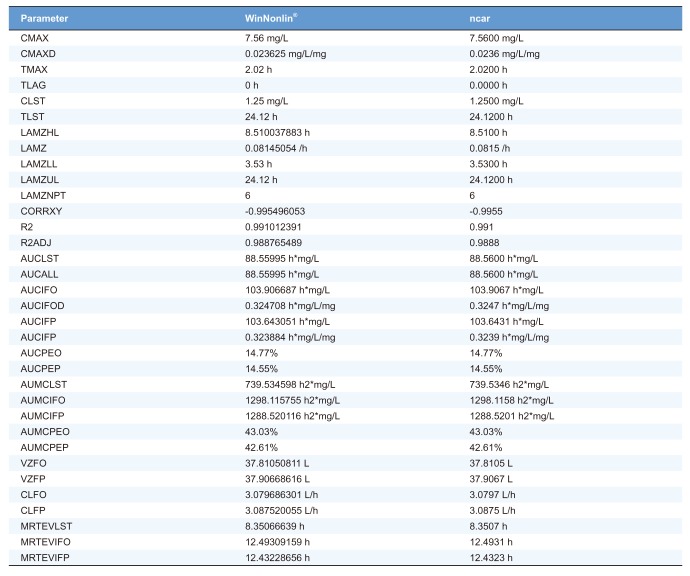
Validation of NCA results between R packages and WinNonlin®
To demonstrate the accordance of outputs by
ncar package and WinNonlin®, we performed NCA using Theoph dataset obtained from the R software. For comparison of the NCA results, we selected the following conditions: extravascular, linear-up linear-down, and best fit. We found no discrepancy between the two results as shown in
Table 3 (a randomized subject, Subject ID = 8).
In order to further validate these packages, we compared NCA results using Indometh, another available dataset of the R software as well as other datasets of a number of subjects from several phase 1 clinical trials with different dosing routes such as infusion, bolus, and oral route. As a result, we could not find any discrepancy between outputs generated by the R packages and WinNonlin®.
Discussion
We developed two R packages—
NonCompart and
ncar for NCA. Through these packages, we aimed to implement the following functionalities for performing NCA: 1) CDISC SDTM terms; 2) automatic slope selection with the same criterion of WinNonlin®; 3) supporting both ‘linear-up linear-down’ and ‘linear-up log-down’ method; and 4) interval (partial) AUCs with ‘linear’ or ‘log’ interpolation method. These packages are convenient and efficient because they enable preparation of data and NCA as well as generation of reports including plots together in R software. As shown in
Figure 2B, the NCA plot allows for automatic slope selection, however, it is not possible to manually choose the points used for calculating terminal slope. In addition, any error or change can easily be fixed, and users may choose calculation methods between linear and logarithmic, which support ‘linear-up linear-down’ and ‘linear-up log-down’ method, respectively. Our results showed that our R packages meet the aforementioned objectives.
Since the PPTESTCD of SDTM is used in the R packages, it is helpful to construct PP domain. In the present practice, one has to change variables from WinNonlin® one by one, which is an especially difficult task for those without specific knowledge on SDTM. A number of packages can perform NCA, but no package—even commercial softwares—can give outputs in the format of SDTM or receive SDTM-formatted input data. It is important to ensure that the reports are legible to sponsors and regulatory bodies by generating a consistent and systematic result, as well as the exact results of NCA.
As shown in
Table 3, comparison of NCA results obtained by WinNonlin® and
ncar package (including another package) showed no significant discrepancies. These two R packages are fast and easy-to-use tool-set that can successfully perform NCA with concentration–time data. Specifically, the
ncar package can produce a comprehensive set of graphical and tabular outputs that summarize the NCA results, which is a complete report in pdf or rtf format. Our two newly-developed packages are free and open-source, so they can be used to develop other useful packages as well. We hope that
NonCompart and
ncar packages will enable researchers to easily perform NCA, and contribute to facilitation of drug discovery process.
Acknowledgements
This research was supported by the EDISON (EDucation-research Integration through Simulation On the Net) Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Grant number: 016M3C1A6936614). We thank Dr. Joon Seo Lim from the Scientific Publications Team at Asan Medical Center for his editorial assistance in preparing this manuscript.
References
1. Acharya C, Hooker AC, Turkyilmaz GY, Jonsson S, Karlsson MO. A diagnostic tool for population models using non-compartmental analysis: The ncappc package for R. Comput Methods Programs Biomed. 2016; 127:83–93. DOI:
10.1016/j.cmpb.2016.01.013. PMID:
27000291.

2. Gabrielsson J, Weiner D. Non-compartmental analysis. Methods Mol Biol. 2012; 929:377. PMID:
23007438.

6. Kim MG, Yim DS, Bae KS. R-based reproduction of the estimation process hidden behind NONMEN Part 1: first-order approximation method. Transl Clin Pharmacol. 2015; 23:1–7.
Figure 1
Example of NonCompart package functions. Example by tblNCA.
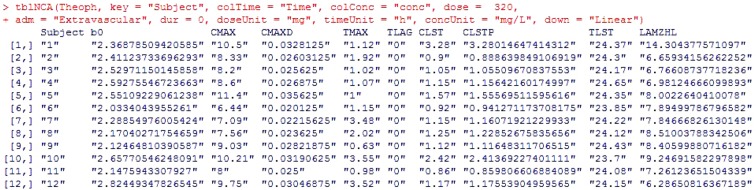
Figure 2
Examples of ncar package functions. (A) Example of a NCA report in pdf format by pdfNCA, (B) Individual concentration-time plots (left: linear scale, right: log-linear scale).
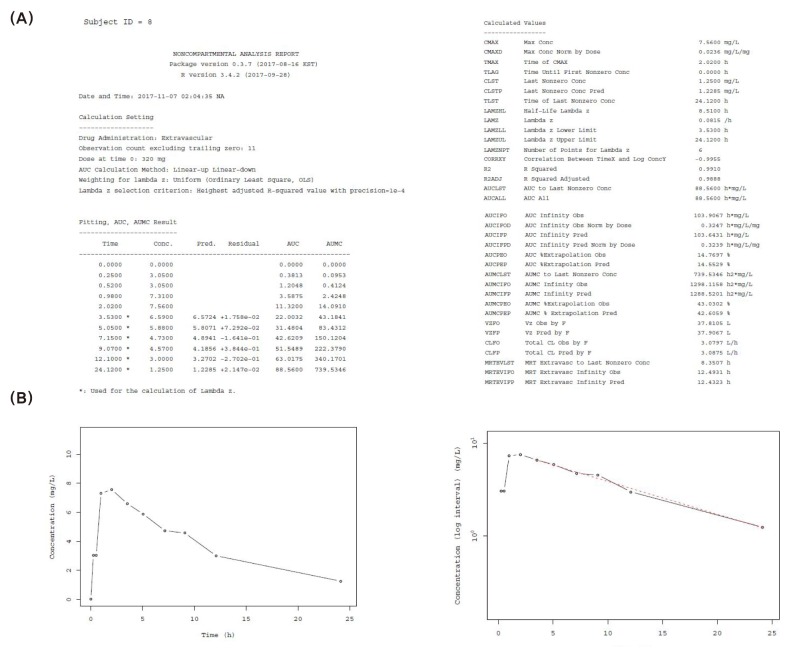
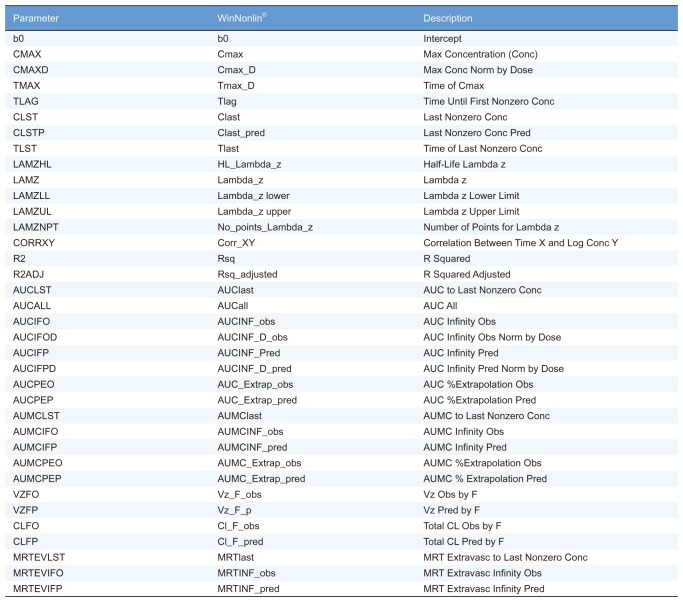
Table 1
Description of PK parameters of WinNonlin® and the R packages
|
Parameter |
WinNonlin® |
Description |
|
b0 |
b0 |
Intercept |
|
CMAX |
Cmax |
Max Concentration (Conc) |
|
CMAXD |
Cmax_D |
Max Conc Norm by Dose |
|
TMAX |
Tmax_D |
Time of Cmax |
|
TLAG |
Tlag |
Time Until First Nonzero Conc |
|
CLST |
Clast |
Last Nonzero Conc |
|
CLSTP |
Clast_pred |
Last Nonzero Conc Pred |
|
TLST |
Tlast |
Time of Last Nonzero Conc |
|
LAMZHL |
HL_Lambda_z |
Half-Life Lambda z |
|
LAMZ |
Lambda_z |
Lambda z |
|
LAMZLL |
Lambda_z lower |
Lambda z Lower Limit |
|
LAMZUL |
Lambda_z upper |
Lambda z Upper Limit |
|
LAMZNPT |
No_points_Lambda_z |
Number of Points for Lambda z |
|
CORRXY |
Corr_XY |
Correlation Between Time X and Log Conc Y |
|
R2 |
Rsq |
R Squared |
|
R2ADJ |
Rsq_adjusted |
R Squared Adjusted |
|
AUCLST |
AUClast |
AUC to Last Nonzero Conc |
|
AUCALL |
AUCall |
AUC All |
|
AUCIFO |
AUCINF_obs |
AUC Infinity Obs |
|
AUCIFOD |
AUCINF_D_obs |
AUC Infinity Obs Norm by Dose |
|
AUCIFP |
AUCINF_Pred |
AUC Infinity Pred |
|
AUCIFPD |
AUCINF_D_pred |
AUC Infinity Pred Norm by Dose |
|
AUCPEO |
AUC_Extrap_obs |
AUC %Extrapolation Obs |
|
AUCPEP |
AUC_Extrap_pred |
AUC %Extrapolation Pred |
|
AUMCLST |
AUMClast |
AUMC to Last Nonzero Conc |
|
AUMCIFO |
AUMCINF_obs |
AUMC Infinity Obs |
|
AUMCIFP |
AUMCINF_pred |
AUMC Infinity Pred |
|
AUMCPEO |
AUMC_Extrap_obs |
AUMC %Extrapolation Obs |
|
AUMCPEP |
AUMC_Extrap_pred |
AUMC % Extrapolation Pred |
|
VZFO |
Vz_F_obs |
Vz Obs by F |
|
VZFP |
Vz_F_p |
Vz Pred by F |
|
CLFO |
Cl_F_obs |
Total CL Obs by F |
|
CLFP |
Cl_F_pred |
Total CL Pred by F |
|
MRTEVLST |
MRTlast |
MRT Extravasc to Last Nonzero Conc |
|
MRTEVIFO |
MRTINF_obs |
MRT Extravasc Infinity Obs |
|
MRTEVIFP |
MRTINF_pred |
MRT Extravasc Infinity Pred |
Table 2
Part of Theoph dataset with information on key (subject), time, and concentration
|
Subject |
Weight (kg) |
Dose (mg) |
Time (h) |
Concentration (mg/ml) |
|
8 |
70.5 |
4.53 |
0 |
0.00 |
|
8 |
70.5 |
4.53 |
0.25 |
3.05 |
|
8 |
70.5 |
4.53 |
0.52 |
3.05 |
|
8 |
70.5 |
4.53 |
0.98 |
7.31 |
|
8 |
70.5 |
4.53 |
2.02 |
7.56 |
|
8 |
70.5 |
4.53 |
3.53 |
6.59 |
|
8 |
70.5 |
4.53 |
5.05 |
5.88 |
|
8 |
70.5 |
4.53 |
7.15 |
4.73 |
|
8 |
70.5 |
4.53 |
9.05 |
4.57 |
|
8 |
70.5 |
4.53 |
12.10 |
3.00 |
|
8 |
70.5 |
4.53 |
24.12 |
1.25 |
Table 3
Comparison of NCA results generated from WinNonlin® and ncar package
|
Parameter |
WinNonlin® |
ncar |
|
CMAX |
7.56 mg/L |
7.5600 mg/L |
|
CMAXD |
0.023625 mg/L/mg |
0.0236 mg/L/mg |
|
TMAX |
2.02 h |
2.0200 h |
|
TLAG |
0 h |
0.0000 h |
|
CLST |
1.25 mg/L |
1.2500 mg/L |
|
TLST |
24.12 h |
24.1200 h |
|
LAMZHL |
8.510037883 h |
8.5100 h |
|
LAMZ |
0.08145054 /h |
0.0815 /h |
|
LAMZLL |
3.53 h |
3.5300 h |
|
LAMZUL |
24.12 h |
24.1200 h |
|
LAMZNPT |
6 |
6 |
|
CORRXY |
−0.995496053 |
−0.9955 |
|
R2 |
0.991012391 |
0.991 |
|
R2ADJ |
0.988765489 |
0.9888 |
|
AUCLST |
88.55995 h*mg/L |
88.5600 h*mg/L |
|
AUCALL |
88.55995 h*mg/L |
88.5600 h*mg/L |
|
AUCIFO |
103.906687 h*mg/L |
103.9067 h*mg/L |
|
AUCIFOD |
0.324708 h*mg/L/mg |
0.3247 h*mg/L/mg |
|
AUCIFP |
103.643051 h*mg/L |
103.6431 h*mg/L |
|
AUCIFP |
0.323884 h*mg/L/mg |
0.3239 h*mg/L/mg |
|
AUCPEO |
14.77% |
14.77% |
|
AUCPEP |
14.55% |
14.55% |
|
AUMCLST |
739.534598 h2*mg/L |
739.5346 h2*mg/L |
|
AUMCIFO |
1298.115755 h2*mg/L |
1298.1158 h2*mg/L |
|
AUMCIFP |
1288.520116 h2*mg/L |
1288.5201 h2*mg/L |
|
AUMCPEO |
43.03% |
43.03% |
|
AUMCPEP |
42.61% |
42.61% |
|
VZFO |
37.81050811 L |
37.8105 L |
|
VZFP |
37.90668616 L |
37.9067 L |
|
CLFO |
3.079686301 L/h |
3.0797 L/h |
|
CLFP |
3.087520055 L/h |
3.0875 L/h |
|
MRTEVLST |
8.35066639 h |
8.3507 h |
|
MRTEVIFO |
12.49309159 h |
12.4931 h |
|
MRTEVIFP |
12.43228656 h |
12.4323 h |




 PDF
PDF ePub
ePub Citation
Citation Print
Print







 XML Download
XML Download