Dear Editor,
Clostridioides difficile infections (CDIs) are antibiotic-associated diseases and have become prevalent healthcare-associated infections [1]. C. difficile is divided into five clades, of which clade 3 (RT023) causes severe infections similar to those seen with the hypervirulent strains of clade 2 (RT027). Shaw, et al. [2] have suggested clade 3 strain characteristics, including a conserved pathogenicity locus, a binary toxin, and an esculinase-negative phenotype. Rapidly and accurately diagnosing CDIs is essential to optimize patient care and ensure proper infection control [3, 4]. Chromogenic media such as ChromID agar have been developed to aid in diagnosis. Most C. difficile strains exhibit esculin hydrolysis, resulting in characteristic black colonies on ChromID agar. The absence of the esculinase enzyme in C. difficile RT023 results in the formation of white colonies on ChromID agar [2, 5]. A recent report highlighted a CDI caused by a strain belonging to the common lineage RT020 (clade 1) that produced white colonies on ChromID agar [6], and 19.5% of strains from environmental samples were esculin hydrolysis-negative [5], suggesting the potential for the widespread dissemination of C. difficile strains that produce white colonies in community-associated CDIs [5, 6]. We report a severe case of infection caused by white colony-producing C. difficile and its genome characteristics.
A 66-year-old man, admitted to a tertiary hospital in Seoul, Korea, in September 2023 with a headache, was diagnosed as having left cerebellar hemorrhage and underwent suboccipital craniectomy. On post-operation day 13, he developed diarrhea (>10 episodes) and fever (37.7°C). Laboratory tests revealed a white blood cell count of 37,900/mm3, acute kidney injury with blood urea nitrogen and creatinine levels of 20.14 mmol/L and 278.52 umol/L, respectively, and hypoalbuminemia (albumin level, 15 g/L). Unenhanced computed tomography revealed a small amount of ascites in the abdominal cavity and diffuse wall thickening extending from the ascending colon to the rectum, accompanied by pericolic infiltration. Nodularity of the colonic wall, a diagnostic indicator of CDI, was noted in the descending colon [7]. Glutamate dehydrogenase antigen and toxin enzyme immunoassays (TcdA and TcdB) using C. Diff Quik Chek Complete (TechLab, Blacksburg, VA, USA) yielded positive results. Toxin B and binary toxin genes were detected in a stool specimen using the Xpert C. difficile assay (Cepheid, Sunnyvale, CA, USA).
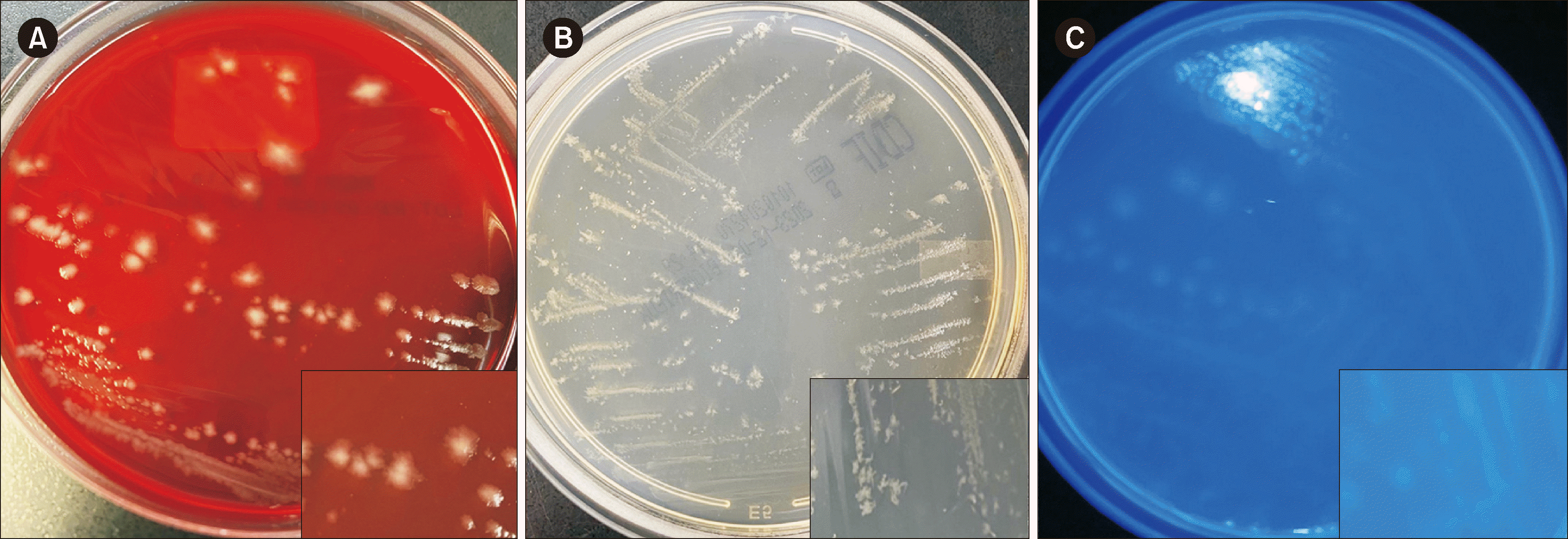
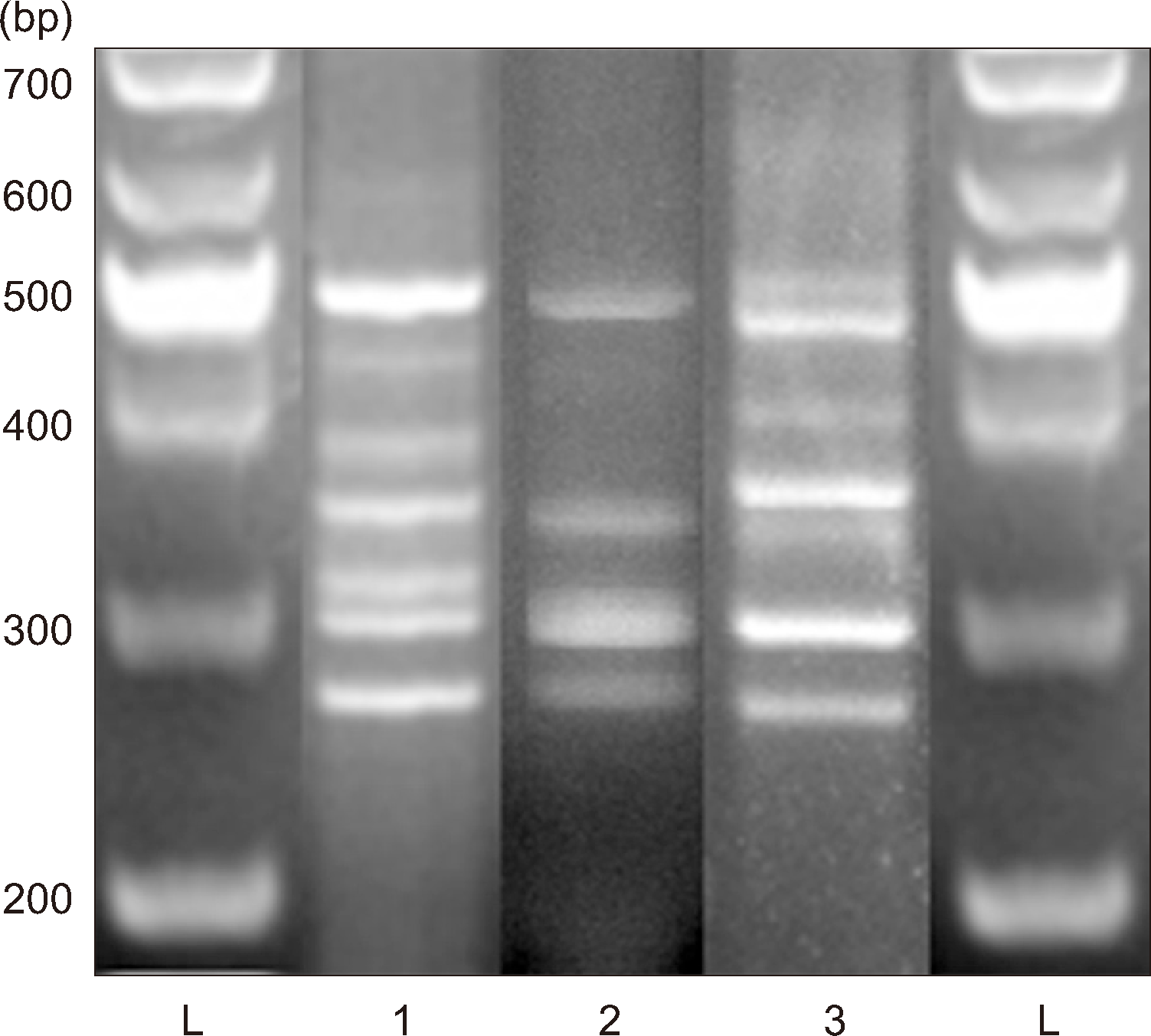
The stool specimen was cultured on ChromID C. difficile agar (bioMérieux, Marcy-l’Étoile, France) under anaerobic conditions for 48 hrs. Grayish irregular colonies were observed on Brucella blood agar (HY1126, Fig. 1A). Although typical black colonies were not observed, flat, irregular, white colonies with a distinctive horse manure odor were observed on ChromID C. difficile agar (bioMérieux) (Fig. 1B). When cultured on CHROM CDIF agar (Asanpharm, Seoul, Korea), the isolate appeared colorless and fluorescent under UV light at 365 nm (Fig. 1C). C. difficile was definitively identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker Biotyper MS; Bruker Daltonics, Bremen, Germany). PCR ribotyping of the HY1126 isolate was conducted using the primers CD1 and CD1445, according to previous protocols [8, 9]. We detected a novel ribotype, which we named RTC41 (Fig. 2).
Whole-genome sequencing using the Illumina system (Illumina, San Diego, CA, USA) was performed at Macrogen (Seoul, Korea). De novo assembly was performed, and the assembly was validated. HY1126 was found to belong to multilocus sequence type (ST) 588, a common ST closely related to ST5 that is a part of clade 3. In strain HY1126, ptsl, which encodes phosphoenolpyruvate protein phosphotransferase, was intact. In RT020 strains, a truncated phosphotransferase enzyme I resulted in impaired esculin transport capability and limited growth [5]. An early stop codon in bglA (1,206 bp) of HY1126, as compared with intact bglA (1,455 bp), determines esculinase-negative phenotypes, such as that in RT023 [2]. HY1126 showed distinctive features among clade 3 strains, including a binary toxin and esculinase-negative phenotype [2]. Strain HY1126 had an intact TcdC gene, responsible for the increased toxin production observed in RT027 strains with an 18-bp deletion in TcdC [10]. The genome sequences were deposited in NCBI GenBank under accession number JAYJMP000000000. This study was approved by the Hanyang University Seoul Hospital Institutional Review Board (approval No. 202311051).
CDI treatment with oral vancomycin and intravenous metronidazole was continued for two weeks, and the patient was discharged with improvement. We report the case of a severe infection caused by C. difficile ST588, a clade 3 strain that produces characteristic white colonies on ChromID agar. The esculinase-negative phenotype may be prevalent among C. difficile isolates, emphasizing the importance of identifying and not disregarding white colonies on ChromID C. difficile agar cultures.
Notes
AUTHOR CONTRIBUTIONS
Lee Y and Park SY designed the study. Lee Y collected and identified the clinical isolates and conducted the molecular analyses. Lee Y, Kim H, and Park SY analyzed the data. Lee Y and Park SY contributed to the writing, editing, and review of the manuscript. All authors participated in the revision and approval of the final manuscript.
References
1. Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, et al. 2020; Trends in U.S. burden of Clostridioides difficile infection and outcomes. N Engl J Med. 382:1320–30. DOI: 10.1056/NEJMoa1910215. PMID: 32242357. PMCID: PMC7861882.

2. Shaw HA, Preston MD, Vendrik KEW, Cairns MD, Browne HP, Stabler RA, et al. 2020; The recent emergence of a highly related virulent Clostridium difficile clade with unique characteristics. Clin Microbiol Infect. 26:492–8. DOI: 10.1016/j.cmi.2019.09.004. PMID: 31525517. PMCID: PMC7167513.
3. Kim MH, Kim YC, Kim JL, Park YS, Kim H. 2022; Description of antibiotic treatment in adults tested for Clostridioides difficile infection: a single-center case-control study. BMC Infect Dis. 22:1–9. DOI: 10.1186/s12879-022-07085-z. PMID: 35093016. PMCID: PMC8801153. PMID: e86375e84c444a2f994e41ad58dbb3d2.
4. Chung HS, Park JS, Shin BM, Yoo HM, Kim H, Cho J, et al. 2022; Nationwide survey for current status of laboratory diagnosis of Clostridioides difficile infection in Korea. J Korean Med Sci. 37:e38. DOI: 10.3346/jkms.2022.37.e38. PMID: 35132844. PMCID: PMC8822111.

5. Shivaperumal N, Knight DR, Imwattana K, Androga GO, Chang BJ, Riley TV. 2022; Esculin hydrolysis negative and TcdA-only producing strains of Clostridium (Clostridioides) difficile from the environment in Western Australia. J Appl Microbiol. 133:1183–96. DOI: 10.1111/jam.15500. PMID: 35184359. PMCID: PMC9544920.
6. Imwattana K, Shivaperumal N, Leepattarakit T, Kiratisin P, Knight DR, Riley TV. 2023; The brief case: a white-colony-producing Clostridioides difficile ribotype 020 strain. J Clin Microbiol. 61:e0089322. DOI: 10.1128/jcm.00893-22. PMID: 37338228. PMCID: PMC10281166.

7. Kirkpatrick ID, Greenberg HM. 2001; Evaluating the CT diagnosis of Clostridium difficile colitis: should CT guide therapy? AJR Am J Roentgenol. 176:635–9. DOI: 10.2214/ajr.176.3.1760635. PMID: 11222194.
8. O'Neill GL, Ogunsola FT, Brazier JS, Duerden BI. 1996; Modification of a PCR ribotyping method for application as a routine typing scheme for Clostridium difficile. Anaerobe. 2:205–9. DOI: 10.1006/anae.1996.0028.
9. Stubbs SL, Brazier JS, O'Neill GL, Duerden BI. 1999; PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 37:461–3. DOI: 10.1128/JCM.37.2.461-463.1999. PMID: 9889244. PMCID: PMC84342.

10. Curry SR, Marsh JW, Muto CA, O'Leary MM, Pasculle AW, Harrison LH. 2007; tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol. 45:215–21. DOI: 10.1128/JCM.01599-06. PMID: 17035492. PMCID: PMC1828959.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download