Abstract
The 2019 novel coronavirus disease (COVID-19) outbreaks that emerged in Wuhan city, Hubei province, have led to a formidable number of confirmed cases that resulted in >5,700 deaths globally, including 143 countries in all 6 continents. The World Health Organization declared a Public Health Emergency of International Concern with a very high level of global risk assessment. Severe acute respiratory syndrome (SARS)-coronavirus-2 (SARS-CoV-2), the agent of COVID-19, has >79% nucleotide sequence homology to SARS-CoV; therefore, both belong to the genus betacoronavirus and subgenus sarbecovirus. The S1 domains of the two appeared to share the cellular receptor ACE2, but revealed a much higher S1-ACE2 binding affinity. As seen in many other human coronaviruses, SARS-CoV-2 also shows respiratory infection, but the basic reproductive number (R0) in transmission and the clinical latency are quite dissimilar from those of SARS- or MERS-CoVs. Many scientists infer that the time point of cross-barrier transfer from bats to mediate animals or to humans should be a rather recent event based on the full-length genome analyses obtained from the very first patients. Copy-choice polymerization, which often leads to a significant genome recombination rate in most coronaviruses, predicts the continued emergence of novel coronaviruses.
REFERENCES
1). Master PS, Perlman S. Coronaviridae. In Fields Virology 6 th Ed. 825–58.
2). Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019; 17:181–92.

3). Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016; 24:490–502.

4). Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020; 63:457–60.

5). Menachery VD, Yount BL Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015; 21:1508–13.

6). Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. Severe acute respiratory syndrome-related coronavirus: the species and its viruses-a statement of the Coronavirus Study Group. BioRxiv. 2020; 1-15.
7). WHO. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
8). Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020; 395:565–74.

10). Luis AD, Hayman DT, O'Shea TJ, Cryan PM, Gilbert AT, Pulliam JR, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc Biol Sci. 2013; 280:20122753.

11). Callaway E, Cyranoski D. What scientists want to know about coronavirus outbreak. Nature (News in focus). 2020; 577:605–7.
12). Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020. pii: eabb2507.

13). Cohen J. Mining coronavirus genomes for clues to the outbreak's origins. 2020 Science.
14). Flint J, Racaniello VR, Rall GF, Skalka AM. Emergence. In Principles of Virology 4 th Ed. 2015. 342–54.
15). Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, Wang M. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020.

16). Al-Tawfig JA, Zumla A, Memish ZA. Travel implications of emerging coronaviruses: SARS and MERS-CoV. Travl Med Infect Dis. 2014; 12:422–8.
17). Noh JW, Yoo KB, Kwon YD, Hong JH, Lee Y, Park K. Effect of information disclosure policy on control of infectious disease: MERS-CoV outbreak in South Korea. Int J Environ Res Public Health. 2020; 17:Pii: E305.

18). Bagamian KH, Towner JS, Kuenzi AJ, Douglass RJ, Rollin PE, Waller LA, et al. Transmission Ecology of Sin Nombre Hantavirus in Naturally Infected North American Deermouse Populations in Outdoor Enclosures. PLoS One. 2012; 7:e47731.

Fig. 1.
Major mechanisms of virus genetic recombination. (A) In nonreplicative recombination, nucleic acid strand breakage and repair permit the recombination of genetic material from different sources into the same viral genome. (B) In replicative recombination or template switching, a polymerase molecule changes template during the process of replicating a nucleic acid strand. If the templates are derived from different sources, then novel genetic material can be introduced into the virus genome. (C) During the process of virus integration and excision from a host genome, viruses can acquire genetic material from the host. (D) Reassortment occurs following coinfection of a host cell by multiple segmented viruses. Replicated genome segments are packaged into procapsids irrespective of the parent of origin. (Adapted from ref. (20), Dennehy J. Evolutionary ecology of virus emergence: Virus emergence. Ann N Y Acad Sci 2016 1389(1) DOI:10.1111/nyas.13304)
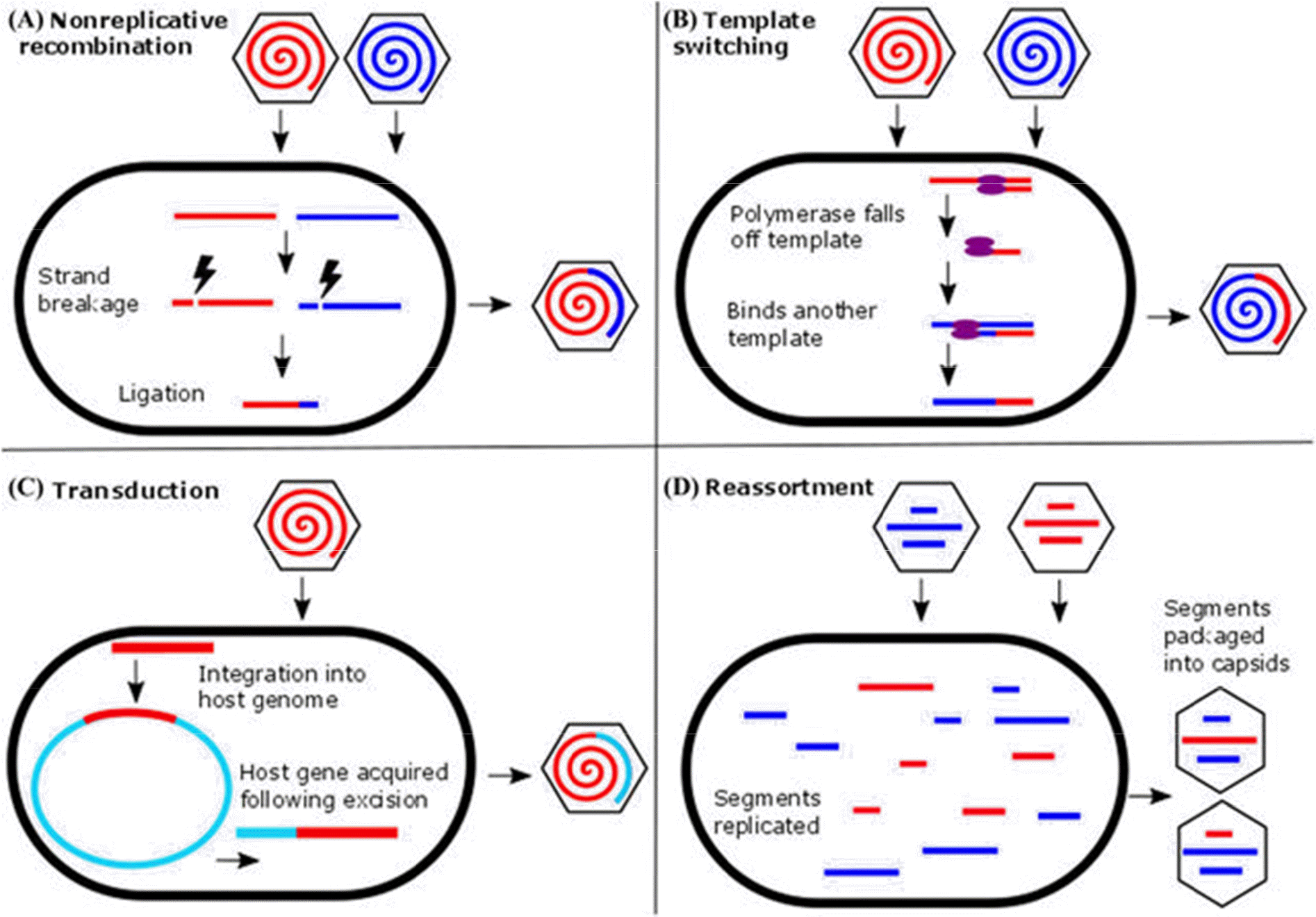
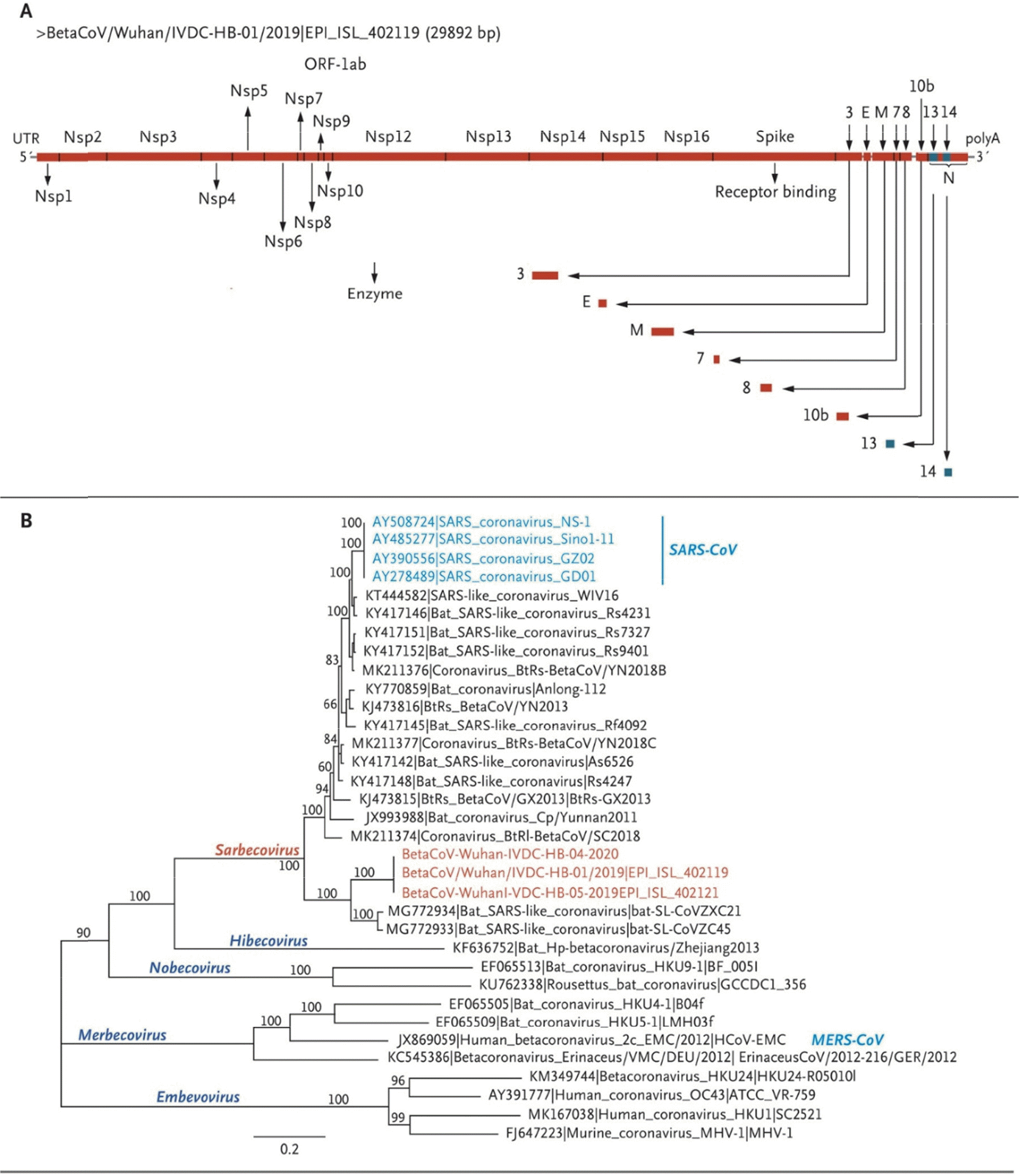
Fig. 2.
Schematic of 2019-nCoV and Phylogenetic Analysis of 2019-nCoV and Other Betacoronavirus Genomes. Shown are a schematic of 2019-nCoV (Panel A) and full-length phylogenetic analysis of 2019-nCoV and other betacoronavirus genomes in the Orthocoronavirinae subfamily (Panel B). (Adapted from ref. (19) Zhu et al., A Novel Coronavirus from Patients with Pneumonia in China, 2019 New Engl J Med 2020;382:727–733)




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download