Dear Editor,
RHD and RHCE, encoding Rhesus proteins, are highly homologous genes located adjacently on the same chromosome (chromosome 1). Therefore, hybrid RHD genes, in which some portions are substituted with the RHCE sequence can change the extracellular loop of the RhD antigen, leading to variable reactivity to anti-D reagents [1].
Partial D phenotypes have historically been classified using epitope studies [2] and, recently, using genetic studies. DBS is a partial D phenotype characterized by c.[676G>C]+[697G>C] (NM_016124.4) and has been named based on its positive reactivity with the D monoclonal antibodies (MoAbs) BS228 and BS233 (Biotest, Dreieich, Germany) [3]. Three DBS subtypes have been reported till date: DBS-0 [1], DBS-1 [34], and DBS-2 [5]. Molecular studies have identified DBS-1 in an Arabian [3] and a Japanese family [4]; however, it is unknown whether DBS-1 correlates with the same hybrid gene of a single evolutionary origin in other, unrelated individuals from different ethnic groups. To the best of our knowledge, this is the first report of a DBS-1 case in a Korean family. This study was approved by the Institutional Review Board of Samsung Medical Center, Seoul, Korea (SMC-2019-11-160), and written informed consent was obtained from the proband and all family members.
The proband was a Korean woman with fibrocystic breast changes, who was admitted to the Samsung Medical Center. D typing using anti-D Bioclone (MAD2 clone; Ortho Clinical Diagnostics, Raritan, NJ, USA) was negative. Weak D testing using anti-D Bioclone and human IgG/IgM monoclonal anti-D (Millipore, Livingston, UK) yielded a result of grade 2+, while the result of partial D testing using D-screen (Diagast, Loos, France) was consistent with DBS-1. The phenotypes of the current case and previously reported DBS cases are summarized in Table 1.
The RhCE phenotype was ccEe (anti-C, -c, -E, and -e antibodies were obtained from Bioclone, Ortho Clinical Diagnostics, Buckinghamshire, UK), and RHD genotyping was carried out according to a previously described method [6]. Exon 5 was not amplified by the primer sets used in this study, but the region from exon 4 to exon 6 was amplified and the PCR product was sequenced using RHCE exon 5-specific primers; the Rhesus box was also PCR-amplified (Fig. 1A). The proband harbored a hybrid RHD-cE(5)-D (DBS/d) allele involving the following amino acid changes: F223V, A226P, E233Q, V238M, V245L, G263R, and K267M. RHD genotypes of other family members are shown in Fig. 1B. The breakpoints were confirmed between exon 4 and exon 6 by intron analysis [34]. The 5′ breakpoint region was the same as that reported by Omi, et al. [4], whereas the 3′ breakpoint region was novel (Fig. 1C).
In various partial D phenotypes, such as DIIIa, DVa, DVI, DAR, DFR, DBT, and DBS, RHD exon 5 is substituted with a part of RHCE exon 5 [7]. Exon 5 is predicted to encode the fourth extracellular loop of the D polypeptide. In DBS, the coexistence of two amino acid changes (A226P and E233Q) caused by c.676G>C and c.697G>C point variants is required for the characteristic phenotype [4]. A226P is observed in the fourth loop of antigen E [4] and is thought to have a considerable effect on D antigen density [1]. E233Q is also observed in the fourth loop as part of Dw (RH23) [8]. DBS-1 and -2, which share F223V, A226P, and E233Q, exhibited different reactivity to the MoAbs P3X249, P3X290, and P3X21211F1 (DBS-1, +/+/− and DBS-2, −/−/trace). This difference might have been caused by the change in the extracellular amino acid, V238M, which is found in DBS-0 and DBS-1, but not in DBS-2. Other amino acid changes observed in DBS-1, including V245L, G263R, and K267M, are located in the intracellular or transmembrane regions; however, they might affect the RhD phenotype.
The genetic basis of the RhD blood group differs across races and ethnicities. For example, the RhD-negative phenotype mainly results from RHD deletion in Caucasians, whereas RHD alterations, such as RHD*D-CE(4-7)-D, are common in Africans [9]. Asians exhibit a high prevalence of c.1227G>A (NM_016124.4), known as “Asia type DEL.” These ethnicity-specific trends are used not only to diagnose RhD variants, but also for transfusion protocols [10]. The 3′ breakpoint observed in our case differed from that found in a Japanese family [4], suggesting that the DBS-1 cases have different genetic origins.
In conclusion, we reported the first case of DBS-1 in a Korean family. To understand the RhD characteristics specific to Korean ethnicity, further evaluation of RhD variants is required.
Notes
References
1. Avent ND, Finning KM, Liu W, Scott ML. Molecular biology of partial D phenotypes. Transfus Clin Biol. 1996; 3:511–516. PMID: 9018818.
2. Flegel WA, Von Zabern I, Doescher A, Wagner FF, Vytisková J, Písačka M. DCS-1, DCS-2, and DFV share amino acid substitutions at the extracellular RhD protein vestibule. Transfusion. 2008; 48:25–33. PMID: 17900276.
3. Wagner FF, Ernst M, Sonneborn HH, Flegel WA. A DV-like phenotype is obliterated by A226P in the partial D DBS. Transfusion. 2001; 41:1052–1058. PMID: 11493738.
4. Omi T, Takahashi J, Seno T, Tanaka M, Hirayama F, Matsuo M, et al. Isolation, characterization, and family study of DTI, a novel partial D phenotype affecting the fourth external loop of D polypeptides. Transfusion. 2002; 42:481–489. PMID: 12076297.
5. Ye L, Wang P, Gao H, Zhang J, Wang C, Li Q, et al. Partial D phenotypes and genotypes in the Chinese population. Transfusion. 2012; 52:241–246. PMID: 21790636.
6. Fasano RM, Monaco A, Meier ER, Pary P, Lee-Stroka AH, Otridge J, et al. RH genotyping in a sickle cell disease patient contributing to hematopoietic stem cell transplantation donor selection and management. Blood. 2010; 116:2836–2838. PMID: 20644109.
7. Wagner F. The Human RhesusBase, version 2.4. Updated on Dec 2018. http://www.rhesusbase.info.
8. Omi T, Okuda H, Iwamoto S, Kajii E, Takahashi J, Tanaka M, et al. Detection of Rh23 in the partial D phenotype associated with the DVa category. Transfusion. 2000; 40:256–257. PMID: 10686014.
9. Wheeler MM, Lannert KW, Huston H, Fletcher SN, Harris S, Teramura G, et al. Genomic characterization of the RH locus detects complex and novel structural variation in multi-ethnic cohorts. Genet Med. 2019; 21:477. PMID: 29955105.
10. Choi S, Chun S, Seo JY, Yang JH, Cho D. Planned transfusion of D-positive blood components in an Asia type DEL patient: proposed modification of the Korean National Guidelines for Blood Transfusion. Ann Lab Med. 2019; 39:102–104. PMID: 30215239.
Fig. 1
Results of the genetic analysis of the proband and her family members. (A) Long-range PCR with primers located in non-Rhesus box sequences. A 2,778-bp fragment was amplified by PCR, indicating the presence of a hybrid RHD gene (lanes I-1, II-1, and II-2). (B) Pedigree, Rh phenotypes, and RHD genotypes of the Korean DBS-1 family. The genotypes and phenotypes of the DBS-1 family were determined using combined data from sequencing analysis, hybrid Rhesus box PCR, and serological analysis. Black circles indicate the DBS-1 phenotype. The proband is indicated by a black arrow. Total RHD deletion is denoted as “d” in the genotype. (C) Part of the RHD nucleotide sequence in DBS reported by Wagner, et al. [3] and this case. In both cases, the 5′ breakpoint region was located between the RHD-specific c.642-249T and the first RHCE-specific nucleotide, c.667G (blue arrow). The 3′ breakpoint region, located between the last RHCE-specific nucleotide and the first RHD-specific nucleotide of intron 5, differed for each case; it was located between c.800 and c.801+101 in the case reported by Wagner, et al. [3] and between c.801+1463 and c.801+1505 in the current case (white arrow).
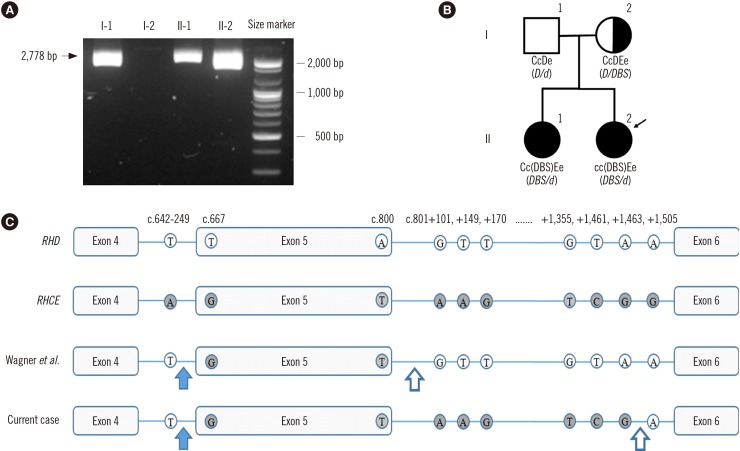
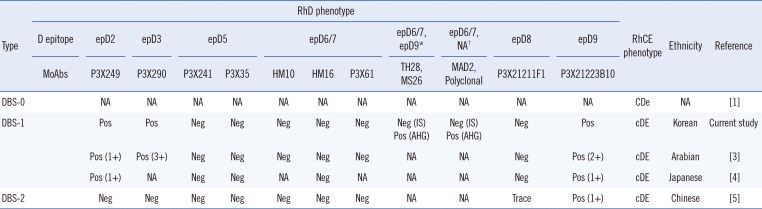
Table 1
Comparison of serologic characteristics based on analysis using MoAbs between previous DBS cases and the present DBS case. Partial D testing was performed using D-screen (Diagast, Loos, France)
| Type | RhD phenotype | RhCE phenotype | Ethnicity | Reference | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D epitope | epD2 | epD3 | epD5 | epD6/7 | epD6/7, epD9* | epD6/7, NA† | epD8 | epD9 | |||||||
| MoAbs | P3X249 | P3X290 | P3X241 | P3X35 | HM10 | HM16 | P3X61 | TH28, MS26 | MAD2, Polyclonal | P3X21211F1 | P3X21223B10 | ||||
| DBS-0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | CDe | NA | [1] | |
| DBS-1 | Pos | Pos | Neg | Neg | Neg | Neg | Neg | Neg (IS) | Neg (IS) | Neg | Pos | cDE | Korean | Current study | |
| Pos (AHG) | Pos (AHG) | ||||||||||||||
| Pos (1+) | Pos (3+) | Neg | Neg | Neg | Neg | Neg | NA | NA | Neg | Pos (2+) | cDE | Arabian | [3] | ||
| Pos (1+) | NA | Neg | Neg | NA | Neg | NA | NA | NA | Neg | Pos (1+) | cDE | Japanese | [4] | ||
| DBS-2 | Neg | Neg | Neg | Neg | Neg | Neg | Neg | NA | NA | Trace | Pos (1+) | cDE | Chinese | [5] | |
*The results using human IgG/IgM monoclonal anti-D (Millipore, Livingston, UK); †The results using anti-D Bioclone (MAD2 clone; Ortho Clinical Diagnostics, Raritan, NJ, USA).
Abbreviations: ep, epitope; MoAbs, monoclonal antibodies; IS, immediate spin; AHG, antihuman globulin; Pos, positive; Neg, negative; NA, not available.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download