Experimental
General experimental procedures
Optical rotations were measured on a Jasco P-2000 polarimeter using methanol solvent. Ultraviolet (UV) spectra were recorded with a Shimadzu UV-1601 UV-vis spectrometer. High Resolution ESI Mass Spectrometer data were obtained with a Waters SYNAPT G2 mass spectrometer. Infrared (IR) spectra were recorded on a Bruker IFS-66/S Fourier-transform IR spectrometer. NMR spectra were recorded on Varian UNITY INOVA 500 NMR spectrometer at 500 MHz (1H) and 125 MHz (13C), and Bruker AVANCE III 700 NMR spectrometer at 700 MHz (1H) and 175 MHz (I. balsamina). Preparative HPLC was performed using a Gilson 321 pump with a Shodex Refractive Index Detector, YMC-Triart C18 5 µm column (250 × 10 mm) and HAISIL 100 silica 5 µm column (250 × 10 mm). Silica gel 60 (Merck, 70 – 230 mesh and 230 – 400 mesh) and RP-C18 silica gel (Merck, 230 – 400 mesh) was used for column chromatography. LPLC was performed over a LiChroprep Lobar-A silica 60 column (Merck, 240 mm × 10 mm i.d.) equipped with a FMI QSY-0 pump. Merck precoated silica gel F254 plates and reversed-phase (RP)-18 F254s plates (Merck) were used for thin-layer chromatography (TLC). Spots were detected on TLC under UV light or by heating after spraying with anisaldehyde-sulfuric acid.
Plant materials
The dried stems of I. balsamina (2 kg) were collected at Asan in Chungcheongnam-Do, Korea, in August 2014. The plants were identified by one of the authors (K.R.L.). A voucher specimen (SKKU-NPL-1425) of the plant was deposited in the herbarium of the School of Pharmacy at Sungkyunkwan University, Suwon, Korea.
Extraction and isolation
The dried stems of I. balsamina (2 kg) were extracted with 80% MeOH at room temperature. The filtrate was evaporated in vacuo to yield MeOH extract (294 g), which was suspended in distilled H2O (2.4 L) and then successively partitioned with hexane, CHCl3, EtOAc and n-BuOH, yielding 20, 11, 6, and 28 g, respectively. The EtOAc fraction (5 g) was separated over a silica gel column with a solvent system of CHCl3/MeOH/water (7:1:0.1) to yielded five sub-fractions (E1–E5). Sub-fraction E1 (1.8 g) was chromatographed on a RP-C18 silica gel column with 30% aqueous MeOH to yield nine sub-fractions (E11–E19). Fraction E12 (430 mg) was purified by RP-C18 semi-prep. HPLC (30% MeOH) to yield 2 (9 mg, tR = 17.1 min). Fraction E2 (1.1 g) was separated by RP-C18 silica gel column with 30% aqueous MeOH to give nine sub-fractions (E21–E29). Sub-fraction E23 (161 mg) was purified using RP-C18 semi-prep. HPLC (30% MeOH) to yield 1 (20 mg, tR = 15.1 min), 6 (24 mg, tR = 21.1 min), and 3 (7 mg, tR = 22.4 min). E3 (557 mg) was subjected to RP-C18 silica gel column with 45% aqueous MeOH to give five sub-fractions (E31–E35). Fraction E31 (148 mg) was purified by RP-C18 semi-prep. HPLC (30% CH3CN) to yield 4 (100 mg, tR = 11.1 min). Fraction E5 (235 mg) was purified using RP-C18 semi-prep. HPLC (20% CH3CN) to yield 5 (32 mg, tR = 21.2 min). The hexane soluble fraction was chromatographed on a silica gel column with hexane/EtOAc (20:1 to 1:1) to give eight sub-fractions (H1–H8). Fraction H2 (546 mg) was subjected to using a RP-C18 silica gel column with 95% aqueous MeOH to yield 11 (9 mg, tR = 34.4 min). Fraction H3 (918 mg) was separated to RP-C18 silica gel column with 90% aqueous MeOH to yield eight sub-fractions (H31–H38). Subfraction H32 (85 mg) was purified using RP-C18 semi-prep. HPLC (90% MeOH) to yield 7 (9 mg, tR = 36.9 min). Fraction H36 (117 mg) was purified by semi-prep. HPLC (CHCl3/MeOH 120:1) to give 12 (88 mg, tR = 11.0 min). Fraction H5 (2.4 g) was chromatographed on a RP-C18 silica gel column (95% aqueous MeOH) to give twelve sub-fractions (H51–H512). Subfraction H56 (89 mg) was purified using RP-C18 semi-prep. HPLC (95% MeOH) to yield 8 (4 mg, tR = 17.8 min). Fraction H57 (169 mg) was purified by semi-prep. HPLC (hexane/EtOAc 3:1) to yield 9 (3 mg, tR = 15.3 min). Fraction H512 (112 mg) was purified using semi-prep. HPLC (hexane/EtOAc 3:1) to give 10 (7 mg, tR = 8.8min).
1β,2α,4β-Triol-1,2,3,4-tetrahydronaphthalene (1)
Colorless gum. :

: −35.8 (
c 0.10, MeOH); IR (KBr) ν
max cm
−1: 3348, 2938, 2836, 1448, 1412, 1055, 1025; UV (MeOH) λ
max (log ε) 201 (0.75), 213 (0.45), 217 (0.38), 262 (0.02) nm;
1H and
I. balsamina NMR : see
Table 1; HRESIMS
m/z 181.0865 [M+H]
+; (calcd. for C
10H
13O
3, 181.0865).
1β,2β,4β-Triol-1,2,3,4-tetrahydronaphthalene (2)
Colorless gum.

: −57.8 (
c 0.11, MeOH); IR (KBr) ν
max cm
−1: 3340, 2944, 2832, 1460, 1408, 1054, 1021; UV (MeOH) λ
max (log ε) 203 (1.28), 211 (1.11), 215 (0.92), 256 (0.04) nm;
1H and
I. balsamina NMR : see
Table 1.
(7R,8S)-Dihydrodehydrodiconiferyl alcohol-9-β-O-D-glucopyranoside (3)
White gum; 1H NMR (CD3OD, 500 MHz): δ 6.99 (1H, d, J = 1.9 Hz, H-2), 6.85 (1H, dd, J = 8.1, 1.9 Hz, H-6), 6.79 (1H, s, H-6′), 6.75 (1H, d, J = 8.1 Hz, H-5), 6.72 (1H, s, H-2′), 5.58 (1H, d, J = 6.4 Hz, H-7), 4.35 (1H, d, J = 7.8 Hz, H-1), 3.85 (3H, s, 3′-OCH3), 3.82 (3H, s, 3-OCH3), 3.56 (2H, t, J = 6.5Hz, H-9′), 2.62 (2H, m, H-7′), 1.81 (2H, tt, J = 13.1, 6.5 Hz, H-8′); I. balsamina NMR (CD3OD, 125 MHz): δ 147.6 (C-3), 146.1 (C-4), 146.0 (C-4′), 143.8 (C-3′), 135.6 (C-1′), 133.2 (C-1), 128.3 (C-5′), 118.4 (C-6), 116.8 (C-6′), 114.6 (C-5), 112.8 (C-2′), 109.4 (C-2), 102.8 (C-1), 87.8 (C-7), 76.8 (C-5), 76.7 (C-3), 73.7 (C-2), 70.9 (C-9), 70.2 (C-4), 61.3 (C-6), 60.8 (C-9′), 55.4 (3′-OCH3), 55.1 (3-OCH3), 51.5 (C-8), 34.4 (C-8′), 31.5 (C-7′).
Kaempferol-3-O-β-D-glucopyranoside (4)
Yellow needles; 1H NMR (CD3OD, 500 MHz): δ 8.08 (2H, d, J = 8.8 Hz, H-2′, 6′), 6.91 (2H, d, J = 8.8 Hz, H-3′, 5′), 6.43 (1H, s, H-8), 6.23 (1H, s, H-6), 5.28 (1H, d, J = 7.4 Hz, H-1); I. balsamina NMR (CD3OD, 125 MHz): δ 178.1 (C-4), 164.7 (C-7), 161.7 (C-5), 160.1 (C-4′), 157.7 (C-9), 157.1 (C-2), 134.0 (C-3), 130.8 (C-2′, 6′), 121.4 (C-1′), 114.7 (C-3′, 5′), 104.3 (C-10), 102.7 (C-1), 98.5 (C-6), 93.6 (C-8), 77.2 (C-5), 76.7 (C-3), 74.3 (C-2), 70.0 (C-4), 61.2 (C-6).
Nicotiflorin (5)
Yellow needles; 1H NMR (CD3OD, 500 MHz): δ 8.09 (2H, d, J = 8.8 Hz, H-2′, 6′), 6.91 (2H, d, J = 8.8 Hz, H-3′, 5′), 6.42 (1H, s, H-8), 6.23 (1H, d, J = 1.8 Hz, H-6), 5.15 (1H, d, J = 7.4 Hz, H-1), 4.54 (1H, d, J = 0.9 Hz, H-1‴), 3.66 (1H, m, H-2‴), 3.55 (1H, dd, J = 9.5, 3.3 Hz, H-3‴), 1.14 (3H, d, J = 6.2 Hz, H-6‴); I. balsamina NMR (CD3OD, 125 MHz): δ 178.0 (C-4), 164.8 (C-7), 161.6 (C-5), 160.1 (C-4′), 158.0 (C-9), 157.2 (C-2), 134.1 (C-3), 130.1 (C-2′, 6′), 121.4 (C-1′), 114.7 (C-3′, 5′), 104.2 (C-10), 103.2 (C-1″), 101.0 (C-1‴), 98.6 (C-6), 93.5 (C-8), 76.8 (C-3″), 75.8 (C-5″), 74.4 (C-2″), 72.5 (C-4‴), 70.9 (C-2‴), 70.7 (C-3‴), 70.0 (C-4″), 68.3 (C-5‴), 67.2 (C-6″), 16.5 (C-6‴).
p-Hydroxybenzoic acid (6)
White gum; 1H NMR (CD3OD, 500 MHz): δ 7.90 (2H, d, J = 8.8 Hz, H-2′, 6′), 6.84 (2H, d, J = 8.8 Hz, H-3′, 5′); I. balsamina NMR (CD3OD, 125 MHz): δ 170.1 (C-7), 163.3 (C-4), 133.0 (C-2, 6), 122.8 (C-1), 116.0 (C-3, 5).
β-Amyrin (7)
Colorless gum; 1H NMR (CDCl3, 500 MHz): δ 5.19 (1H, t, J = 3.6 Hz, H-12), 3.22 (1H, dd, J = 11.0, 4.6 Hz, H-3), 1.14 (3H, s, H-27), 1.01 (3H, s, H-26), 0.97 (3H, s, H-23), 0.94 (3H, s, H-25), 0.88 (6H, s, H-29, 30), 0.84 (3H, s, H-28), 0.80 (3H, s, H-24); I. balsamina NMR (CDCl3, 175 MHz): δ 145.4 (C-13), 121.9 (C-12), 79.3 (C-3), 55.4 (C-5), 47.8 (C-9), 47.4 (C-18), 47.0 (C-19), 41.9 (C-14), 40.0 (C-8), 39.0 (C-4), 38.8 (C-1), 37.3 (C-10), 37.2 (C-22), 34.9 (C-21), 33.5 (C-29), 32.8 (C-7), 32.7 (C-17), 31.3 (C-20), 28.6 (C-28), 28.3 (C-23), 27.3 (C-2), 27.1 (C-16), 26.3 (C-15), 26.2 (C-27), 23.9 (C-30), 23.7 (C-11), 18.5 (C-6), 17.0 (C-26), 15.8 (C-25), 15.7 (C-24).
Erythrodiol (8)
Colorless gum; 1H NMR (CDCl3, 500 MHz): δ 5.20 (1H, t, J = 3.5 Hz, H-12), 3.55 (1H, d, J = 10.9 Hz, H-28a), 3.22 (1H, d, J = 10.9 Hz, H-28b), 3.22 (1H, d, J = 10.9 Hz, H-3), 1.17 (3H, s, H-27), 1.00 (3H, s, H-23), 0.95 (3H, s, H-26), 0.94 (3H, s, H-25), 0.89 (3H, s, H-29), 0.88 (3H, s, H-30), 0.79 (3H, s, H-24); I. balsamina NMR (CDCl3, 175 MHz): δ 144.2 (C-13), 122.4 (C-12), 79.0 (C-3), 69.7 (C-28), 55.2 (C-5), 47.6 (C-9), 46.5 (C-19), 42.4 (C-18), 41.7 (C-14), 39.8 (C-8), 38.9 (C-4), 38.6 (C-1), 37.0 (C-10, 17), 34.1 (C-21), 33.2 (C-29), 32.6 (C-7), 31.0 (C-20), 30.9 (C-22), 28.0 (C-23), 27.2 (C-2), 25.9 (C-27), 25.6 (C-15), 23.6 (C-11), 23.5 (C-30), 22.0 (C-16), 18.3 (C-6), 16.7 (C-26), 15.6 (C-24), 15.5 (C-25).
α-Spinasterol (9)
White needles; 1H NMR (CDCl3, 500 MHz): δ 5.16 (1H, dd, J = 15.7, 8.7 Hz, H-22), 5.03 (1H, dd, J = 15.2, 8.6 Hz, H-23), 3.60 (1H, tt, J = 10.9, 4.5 Hz, H-3), 1.03 (3H, d, J = 6.6 Hz, H-3), 0.85 (3H, d, J = 6.5 Hz, H-26), 0.56 (3H, s, H-18); I. balsamina NMR (CDCl3, 175 MHz): δ 139.8 (C-8), 139.4 (C-22), 129.7 (C-23), 117.7 (C-7), 71.3 (C-3), 56.1 (C-17), 55.4 (C-14), 51.5 (C-24), 49.7 (C-9), 43.5 (C-13), 41.0 (C-20), 40.5 (C-5), 39.7 (C-12), 38.2 (C-4), 37.4 (C-1), 34.4 (C-10), 32.1 (C-25), 31.7 (C-2), 29.9 (C-6), 28.7 (C-16), 25.6 (C-28), 23.2 (C-15), 21.8 (C-11), 21.6 (C-26), 21.3 (C-21), 19.2 (C-27), 13.2 (C-19), 12.5 (C-29), 12.3 (C-18).
29-Nor-20-oxolupeol (10)
White gum; 1H NMR (CDCl3, 500 MHz): δ 3.19 (1H, dd, J = 11.4, 4.8 Hz, H-3), 2.58 (1H, td, J = 11.3, 6.0 Hz, H-19), 2.15 (3H, s, H-30), 1.02 (3H, s, H-26), 0.97 (6H, s, H-23, 27), 0.83 (3H, s, H-25), 0.78 (3H, s, H-28), 0.76 (3H, s, H-24); I. balsamina NMR (CDCl3, 175 MHz): δ 213.1 (C-20), 79.2 (C-3), 55.5 (C-5), 52.9 (C-19), 50.5 (C-9), 49.9 (C-18), 43.3 (C-14), 42.9 (C-17), 41.0 (C-8), 40.1 (C-22), 39.1 (C-4), 38.9 (C-1), 37.4 (C-10), 37.3 (C-13), 35.2 (C-16), 34.4 (C-7), 29.4 (C-30), 28.2 (C-23), 27.9 (C-21), 27.6 (C-15), 27.5 (C-2), 27.4 (C-12), 21.1 (C-11), 18.5 (C-6), 18.2 (C-28), 16.3 (C-25), 16.1 (C-26), 15.6 (C-24), 14.7 (C-27).
Lupenone (11)
Colorless gum; 1H NMR (CDCl3, 500 MHz): δ 4.69 (1H, d, J = 2.4 Hz, H-29a), 4.57 (1H, dd, J = 2.4, 1.4 Hz, H-29b), 1.69 (3H, brs, H-30), 1.07 (6H, s, H-24, 26), 1.03 (3H, s, H-23), 0.96 (3H, s, H-27), 0.93 (3H, s, H-25), 0.80 (3H, s, H-28); I. balsamina NMR (CDCl3, 175 MHz): δ 218.2 (C-3), 150.9 (C-20), 109.4 (C-29), 54.9 (C-5), 49.8 (C-9), 48.2 (C-18), 47.9 (C-19), 47.3 (C-4), 43.0 (C-14), 42.9 (C-17), 40.8 (C-8), 40.0 (C-22), 39.6 (C-2), 38.2 (C-13), 36.9 (C-10), 35.5 (C-16), 34.1 (C-1), 33.6 (C-7), 29.8 (C-21), 27.4 (C-15), 26.6 (C-23), 25.1 (C-12), 21.5 (C-11), 21.0 (C-24), 19.7 (C-6), 19.3 (C-30), 18.0 (C-28), 16.0 (C-25), 15.8 (C-26), 14.5 (C-27).
Lupeol (12)
White gum; 1H NMR (CDCl3, 500 MHz): δ 4.69 (1H, d, J = 2.3 Hz, H-29a), 4.57 (1H, dd, J = 2.4, 1.4 Hz, H-29b), 3.19 (1H, dd, J = 11.4, 4.9 Hz, H-3), 2.37 (1H, td, J = 11.1, 5.8 Hz, H-19), 1.68 (3H, brs, H-30), 1.03 (3H, s, H-26), 0.97 (3H, s, H-27), 0.95 (3H, s, H-23), 0.83 (3H, s, H-25), 0.79 (3H, s, H-24), 0.76 (3H, s, H-28); I. balsamina NMR (CDCl3, 175 MHz): δ 151.2 (C-20), 109.4 (C-29), 79.1 (C-3), 55.6 (C-5), 50.5 (C-9), 48.7 (C-19), 48.3 (C-18), 43.2 (C-17), 43.1 (C-14), 41.1 (C-8), 40.3 (C-22), 39.0 (C-1, 4), 38.3 (C-13), 37.2 (C-10), 35.6 (C-16), 34.4 (C-7), 30.1 (C-21), 28.4 (C-23), 27.9 (C-15), 27.7 (C-2), 25.4 (C-12), 21.3 (C-11), 19.7(C-30), 18.6 (C-6), 18.3(C-28), 16.5 (C-25), 16.3 (C-26), 15.7 (C-24), 14.7 (C-27).
Measurement of NO Production and Cell Viability
Murine microglial BV-2 cells were plated into a 96-well plate (3 × 104 cells/well). After 24 h, cells were pretreated with samples for 30 min and then stimulated with 100 ng/mL of LPS for another 24 h. Nitrite, a soluble oxidation product of NO, was measured in culture media using the Griess reaction. The supernatant (50 µL) was harvested and mixed with an equal volume of Griess reagent (1% sulfanilamide, 0.1% N-1-naphthylethylenediamine dihydrochloride in 5% phosphoric acid). After 10 min, the absorbance at 540 nm was measured using a microplate reader. Sodium nitrite was used as a standard to calculate the NO2 - concentration. Cell viability was tested by the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide (MTT) assay. NG-Mono-methyl-L-arginine was evaluated as a positive control.
NGF and cell viability assays
C6 glioma cells were used to measure NGF release into the medium. C6 cells were purchased from the Korean Cell Line Bank and maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin in a humidified incubator with 5% CO2. C6 cells were seeded into 24-well plates (1 × 105 cells/well). After 24 h, the cells were treated with DMEM containing 2% FBS and 1% penicillin-streptomycin with 20 µM of each sample for one day. The cell viability of the C6 cells was tested via an MTT assay. 6-shogaol was evaluated as a positive control.
Result and Discussion
The structures of compounds
2 –
12 were determined by comparison of their spectroscopic data with those in the literatures to be 1β,2β,4β-triol-1,2,3,4-tetrahydronaphthalene (
2),
11 (7
R,8
S)-dihydrodehydrodiconiferyl alcohol-9-
O-β-D-glucopyranoside (
3),
12 kaempferol-3-
O-β-D-glucopyranoside (
4),
13 nicotiflorin (
5),
14 p-hydroxybenzoic acid (
6),
15 β-amyrin (
7),
16 erythrodiol (
8),
17 α-spinasterol (
9),
18 29-nor-20-oxolupeol (
10),
19 lupenone (
11),
20 and lupeol (
12)
21 (
Fig. 1).
Compound
1 was obtained as a colorless gum. The molecular formula was determined to be C
10H
12O
3, based on molecular ion peak [M+H]
+ at
m/z 181.0865 (calcd for C
10H
13O
3, 118.0865) in the positive-ion HR-ESI-MS. The IR spectrum of
1 indicated the presence of hydroxy group (3348 cm
−1). The
1H NMR spectrum of
1 showed the presence of a 1,2-disubstituted aromatic ring protons at δ
H 7.56 (1H, d,
J = 7.3 Hz), 7.41 (1H, dd,
J = 7.3, 1.5 Hz), 7.34 (1H, td,
J = 7.3, 1.5 Hz), and 7.29 (1H, td,
J = 7.3, 1.5 Hz), three oxygenated proton signals at δ
H 4.89 (1H, m), 4.41 (1H, d,
J = 7.3 Hz), and 4.09 (1H, ddd,
J = 10.1, 7.3, 3.4 Hz) and one methylene proton signal at δ
H 2.21 (1H, ddd,
J = 13.5, 4.7, 3.4 Hz), and 2.12 (1H, ddd,
J = 13.5, 10.1, 4.7 Hz). The
I. balsamina NMR spectrum displayed 10 carbon signals, including aromatic carbon signals at δ
C 137.6, 137.5, 128.1, 127.6, 127.4, and 127.1, three oxygenated carbon signals at δ
C 73.6, 68.9, and 66.6, one methylene carbon signal at δ
C 37.1. These NMR data were similar to 1α,2α,4β-triol-1,2,3,4-tetrahydronaphthalene, except for chemical shifts and coupling constants of methine protons.
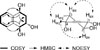
22 The location of hydroxyl groups was confirmed by
1H-
1H COSY correlations and HMBC data (
Fig. 2). The relative configuration of
1 established based on coupling constants (
3J1,2 = 7.3 Hz,
3J2,3 = 10.1, 3.4 Hz, and
3J3,4 = 4.7 Hz) of
1H-NMR spectra, leading oxygenated protons at C-1, C-2, and C-4 to be as axial, axial, and equatorial positions, respectively (
Fig. 2).
2223 In the NOESY spectrum, correlations of H-3
ax with H-1 and H-4 indicated that OH groups at C-1 and C-4 were to be in the same orientation, and cross peak H-2/H-3
eq showed that OH groups at C-1 and C-2 were to be in the opposite orientation (
Fig. 2). Thus, the structure of
1 was established as 1β,2α,4β-triol-1,2,3,4-tetrahydronaphthalene.
The isolated compounds (
1 –
12) were evaluated for their inhibitory effects on NO production in LPS-activated murine microglial cells and their effects on NGF secretion from C6 glioma cells. Among them, compounds
3,
7, and
10 reduced NO levels in LPS-activated murine microglial BV-2 cells with IC
50 values of 26.89, 25.59, and 44.21 µM, respectively. Compounds
1,
5, and
9 upregulated NGF secretion to 153.09 ± 4.66, 156.88 ± 8.86, and 157.34 ± 3.30%, respectively (
Tables 2 and
3).
 : −35.8 (c 0.10, MeOH); IR (KBr) νmax cm−1: 3348, 2938, 2836, 1448, 1412, 1055, 1025; UV (MeOH) λmax (log ε) 201 (0.75), 213 (0.45), 217 (0.38), 262 (0.02) nm; 1H and I. balsamina NMR : see Table 1; HRESIMS m/z 181.0865 [M+H]+; (calcd. for C10H13O3, 181.0865).
: −35.8 (c 0.10, MeOH); IR (KBr) νmax cm−1: 3348, 2938, 2836, 1448, 1412, 1055, 1025; UV (MeOH) λmax (log ε) 201 (0.75), 213 (0.45), 217 (0.38), 262 (0.02) nm; 1H and I. balsamina NMR : see Table 1; HRESIMS m/z 181.0865 [M+H]+; (calcd. for C10H13O3, 181.0865). : −57.8 (c 0.11, MeOH); IR (KBr) νmax cm−1: 3340, 2944, 2832, 1460, 1408, 1054, 1021; UV (MeOH) λmax (log ε) 203 (1.28), 211 (1.11), 215 (0.92), 256 (0.04) nm; 1H and I. balsamina NMR : see Table 1.
: −57.8 (c 0.11, MeOH); IR (KBr) νmax cm−1: 3340, 2944, 2832, 1460, 1408, 1054, 1021; UV (MeOH) λmax (log ε) 203 (1.28), 211 (1.11), 215 (0.92), 256 (0.04) nm; 1H and I. balsamina NMR : see Table 1.



 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download