Abstract
Objective
The objective of this study is to assess whether genetic functional variants of disintegrin and metalloprotease 33 (ADAM33) are associated with susceptibility to systemic lupus erythematosus (SLE) in a Korean population.
Methods
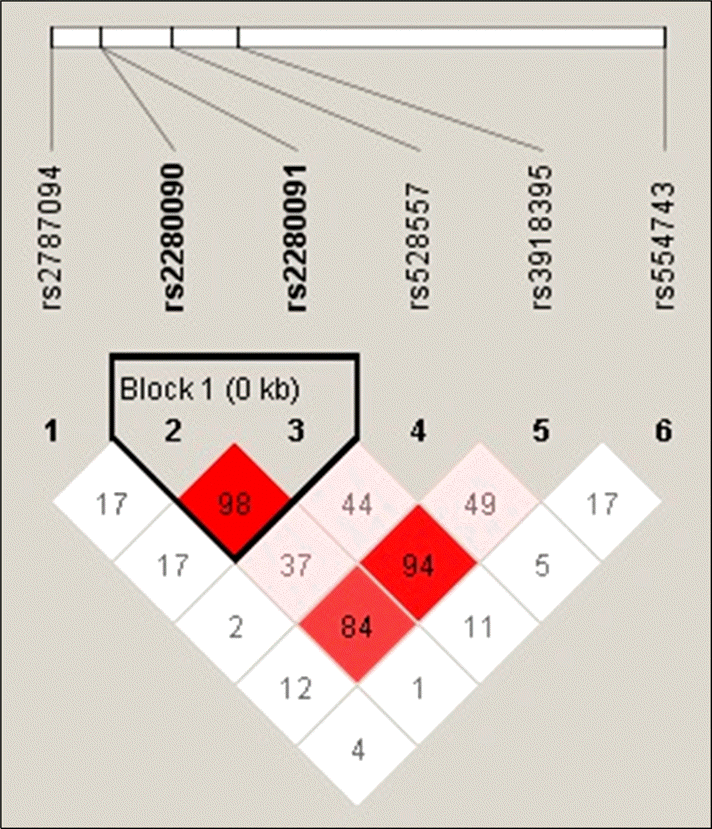
We previously identified 48 single nucleotide polymorphisms (SNPs) in ADAM33. Six SNPs were selected with regard to the linkage disequilibrium pattern. An association study of ADAM33 was conducted in 190 patients with SLE and 469 control subjects. SNPs were genotyped using the TaqMan Real-time polymerase chain reaction method, and haplotype analyses of related variants were performed.
Results
All SNPs were in Hardy-Weinberg equilibrium. Significant associations were found between the ADAM33 polymorphisms and SLE at rs2787094 (adjusted odds ratio [OR] 1.88, 95% confidence interval [CI] 1.00 to 3.54; p< 0.0001). The rs554743 polymorphism was associated with the presence of the immunoglobulin M anti-cardiolipin antibody (adjusted OR 0.29, 95% CI 0.10 to 0.83; p=0.021).
REFERENCES
1. Tan EM. Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv Immunol. 1989; 44:93–151.

2. Tsao BP. Update on human systemic lupus erythematosus genetics. Curr Opin Rheumatol. 2004; 16:513–21.

3. Herrmann M, Voll RE, Kalden JR. Etiopathogenesis of systemic lupus erythematosus. Immunol Today. 2000; 21:424–6.

4. Gunn TM, Azarani A, Kim PH, Hyman RW, Davis RW, Barsh GS. Identification and preliminary characterization of mouse Adam33. BMC Genet. 2002; 3:2.
5. Yoshinaka T, Nishii K, Yamada K, Sawada H, Nishiwaki E, Smith K, et al. Identification and characterization of novel mouse and human ADAM33s with potential metalloprotease activity. Gene. 2002; 282:227–36.

6. Stone AL, Kroeger M, Sang QX. Structure-function analysis of the ADAM family of disintegrin-like and metalloproteinase-containing proteins (review). J Protein Chem. 1999; 18:447–65.
7. Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995; 131:275–8.

9. Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000; 16:83–7.

10. Howard L, Maciewicz RA, Blobel CP. Cloning and characterization of ADAM28: evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem J. 2000; 348:21–7.

11. Shi Z, Xu W, Loechel F, Wewer UM, Murphy LJ. ADAM 12, a disintegrin metalloprotease, interacts with insulin-like growth factor-binding protein-3. J Biol Chem. 2000; 275:18574–80.

12. Wolfsberg TG, Straight PD, Gerena RL, Huovila AP, Primakoff P, Myles DG, et al. ADAM, a widely distributed and developmentally regulated gene family encoding membrane proteins with a disintegrin and metalloprotease domain. Dev Biol. 1995; 169:378–83.
13. Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002; 418:426–30.

14. Sharma N, Tripathi P, Awasthi S. Role of ADAM33 gene and associated single nucleotide polymorphisms in asthma. Allergy Rhinol (Providence). 2011; 2:e63–70.

15. Xiao J, Han J, Wang X, Hua D, Su D, Bao Y, et al. Association of ADAM33 gene with susceptibility to COPD in Tibetan population of China. Mol Biol Rep. 2011; 38:4941–5.

16. Jie Z, Jin M, Cai Y, Bai C, Shen Y, Yuan Z, et al. The effects of Th2 cytokines on the expression of ADAM33 in aller-gen-induced chronic airway inflammation. Respir Physiol Neurobiol. 2009; 168:289–94.

17. Funauchi M, Ikoma S, Enomoto H, Horiuchi A. Decreased Th1-like and increased Th2-like cells in systemic lupus erythematosus. Scand J Rheumatol. 1998; 27:219–24.
18. Llorente L, Richaud-Patin Y, Wijdenes J, Alcocer-Varela J, Maillot MC, Durand-Gasselin I, et al. Spontaneous production of interleukin-10 by B lymphocytes and monocytes in systemic lupus erythematosus. Eur Cytokine Netw. 1993; 4:421–7.
19. Rüger BM, Erb KJ, He Y, Lane JM, Davis PF, Hasan Q. Interleukin-4 transgenic mice develop glomerulosclerosis independent of immunoglobulin deposition. Eur J Immunol. 2000; 30:2698–703.
20. Zhao M, Tang J, Gao F, Wu X, Liang Y, Yin H, et al. Hypomethylation of IL10 and IL13 promoters in CD4+ T cells of patients with systemic lupus erythematosus. J Biomed Biotechnol. 2010; 2010; 931018.
21. Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997; 40:1725.

22. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988; 31:315–24.

23. Chae SC, Yoon KH, Chung HT. Identification of novel polymorphisms in the Adam33 gene. J Hum Genet. 2003; 48:278–81.

24. Lee JH, Park HS, Park SW, Jang AS, Uh ST, Rhim T, et al. ADAM33 polymorphism: association with bronchial hyperresponsiveness in Korean asthmatics. Clin Exp Allergy. 2004; 34:860–5.

25. Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982; 25:1271–7.

26. Priori R, Medda E, Conti F, Cassara EA, Danieli MG, Gerli R, et al. Familial autoimmunity as a risk factor for systemic lupus erythematosus and vice versa: a case-control study. Lupus. 2003; 12:735–40.
27. Schulze-Koops H, Kalden JR. The balance of Th1/Th2 cytokines in rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2001; 15:677–91.

28. Mori M, Morris SC, Orekhova T, Marinaro M, Giannini E, Finkelman FD. IL-4 promotes the migration of circulating B cells to the spleen and increases splenic B cell survival. J Immunol. 2000; 164:5704–12.

29. Dörner T, Jacobi AM, Lee J, Lipsky PE. Abnormalities of B cell subsets in patients with systemic lupus erythematosus. J Immunol Methods. 2011; 363:187–97.

30. Cappione AJ, Pugh-Bernard AE, Anolik JH, Sanz I. Lupus IgG VH4.34 antibodies bind to a 220-kDa glycoform of CD45/B220 on the surface of human B lymphocytes. J Immunol. 2004; 172:4298–307.

31. Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE. Identification and characterization of circulating human transitional B cells. Blood. 2005; 105:4390–8.

Table 1.
Clinical characteristics of the study subjects
Table 2.
Basic information on the genotyped single nucleotide polymorphisms (SNPs) in the ADAM33 gene
| SNP No. | NCBI rs No. | Chromosome position* | e Location | Base change | Amino acid | MAF | HWE‡ | Success rate (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| JPT† | CHB† | SLE | control | ||||||||
| 1 | rs554743 | 3662142 | Intron1 | A> G | 0.43 | 0.39 | 0.403 | 0.382 | 0.602 | 92.39 | |
| 2 | rs3918395 | 3653149 | Intron13 | G> T | 0.13 | 0.09 | 0.082 | 0.073 | 1 | 95.52 | |
| 3 | rs528557 | 3651742 | Exon19 | G> C | Gly717Gly | 0.27 | 0.20 | 0.236 | 0.234 | 0.135 | 92.99 |
| 4 | rs2280091 | 3650234 | Exon20 | T> C | Met764Thr | 0.14 | 0.09 | 0.063 | 0.068 | 1 | 97.01 |
| 5 | rs2280090 | 3650205 | Exon20 | C> T | Pro774Ser | 0.16 | 0.11 | 0.060 | 0.077 | 0.670 | 95.37 |
| 6 | rs2787094 | 3649161 | 3'UTR | C> G | 0.28 | 0.40 | 0.367 | 0.337 | 0.591 | 93.88 | |
CHB: Han Chinese in Bejing, China, JPT: Japanese in Tokyo, Japan; MAF: minor allele frequency, NCBI: National Center fo Biotechnology Information, SLE: systemic lupus erythematosus, UTR: untranslated region.
* SNP position from the NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp).
† MAF from http://browser.1000genomes.org/.
Table 3.
Genotypic and allelic analysis of the ADAM33 gene polymorphisms in the controls and patients with SLE or RA (disease control)
| SNP No. | NCBI rs No. | Genotype | Control | SLE | Adjusted OR (95% CI) | p-value* (control vs. SLE) | p-value* (control vs. RA) |
|---|---|---|---|---|---|---|---|
| 1 | rs554743 | Total (AA, AG, GG) | 429 (100.0) | 190 (100.0) | 487 | ||
| AA | 165 (38.5) | 62 (32.6) | 1.00 (reference) | 0.796 | 0.616 | ||
| AG | 200 (46.6) | 97 (51.1) | 1.13 (0.69∼1.87) | ||||
| GG | 64 (14.9) | 31 (16.3) | 0.93 (0.48∼1.80) | ||||
| AG/GG vs. AA | 264 (61.5) | 128 (67.4) | 1.07 (0.67∼1.72) | 0.768 | 0.650 | ||
| AA/AG vs. GG | 365 (85.1) | 159 (83.7) | 0.87 (0.48∼1.57) | 0.637 | 0.502 | ||
| G allele freq. | 0.382 | 0.403 | 1.09 (0.12∼0.87) | 0.459 | 0.271 | ||
| 2 | rs3918395 | Total (GG, GT, TT) | 462 (100.0) | 178 (100.0) | 485 | ||
| GG | 389 (84.2) | 156 (87.6) | 1.00 (reference) | 0.801 | 1.000 | ||
| GT | 71 (15.4) | 19 (10.7) | 0.83 (0.43∼1.59) | ||||
| TT | 2 (0.4) | 3 (1.7) | 1.41 (0.16∼12.20) | ||||
| GT/TT vs. GG GG/GT vs. TT | 73 (15.8) 460 (99.6) | 22 (12.4) 175 (98.3) | 0.86 (0.46∼1.61) 1.44 (0.17∼12.46) | 0.641 0.740 | 0.990 0.997 | ||
| T allele freq. | 0.074 | 0.082 | 1.12 (0.74∼1.69) | 0.588 | 0.482 | ||
| 3 | rs528557 | Total (GG, GC, CC) | 463 (100.0) | 160 (100.0) | 477 | ||
| GG | 274 (59.2) | 98 (61.3) | 1.00 (reference) | 0.864 | 0.281 | ||
| GC | 156 (33.7) | 52 (32.5) | 0.87 (0.53∼1.43) | ||||
| CC | 33 (7.1) | 10 (6.3) | 0.97 (0.38∼2.45) | ||||
| GC/CC vs. GG | 189 (40.8) | 62 (38.8) | 0.89 (0.56∼1.42) | 0.618 | 0.914 | ||
| GG/GC vs. CC | 430 (92.9) | 150 (93.8) | 1.02 (0.41∼2.54) | 0.969 | 0.122 | ||
| C allele freq. | 0.234 | 0.237 | 1.02 (0.78∼1.32) | 0.913 | 0.810 | ||
| 4 | rs2280091 | Total (AA, AG, GG) | 457 (100.0) | 192 (100.0) | 487 | ||
| TT | 389 (85.1) | 178 (92.7) | 1.00 (reference) | 0.172 | 0.864 | ||
| TC | 66 (14.4) | 13 (6.8) | 0.50 (0.24∼1.05) | ||||
| CC | 2 (0.4) | 1 (0.5) | 0.52 (0.04∼7.72) | ||||
| TC/CC vs. TT | 68 (14.9) | 14 (7.3) | 0.50 (0.25∼1.03) | 0.061 | 0.603 | ||
| TT/TC vs. CC | 389 (85.1) | 191 (99.5) | 2.04 (0.14∼30.29) | 0.604 | 0.985 | ||
| C allele freq. | 0.069 | 0.063 | 0.92 (0.59∼1.43) | 0.702 | 0.282 | ||
| 5 | rs2280090 | Total (AA, AG, GG) | 449 (100.0) | 190 (100.0) | 489 | ||
| CC | 379 (84.4) | 177 (93.2) | 1.00 (reference) | 0.141 | 0.245 | ||
| CT | 68 (15.1) | 12 (6.3) | 0.47 (0.22∼1.01) | ||||
| TT | 2 (0.4) | 1 (0.5) | 1.89 (0.06∼56.43) | ||||
| CT/TT vs. CC | 70 (15.6) | 13 (6.8) | 0.50 (0.24∼1.05) | 0.066 | 0.147 | ||
| CC/CT vs. TT | 447 (99.6) | 189 (99.5) | 2.02 (0.07∼60.49) | 0.684 | 0.250 | ||
| T allele freq. | 0.077 | 0.060 | 0.77 (0.50∼1.19) | 0.235 | 0.348 | ||
| 6 | rs2787094 | Total (CC, CG, GG) | 445 (100.0) | 184 (100.0) | 489 | ||
| CC | 186 (41.8) | 91 (49.5) | 1.00 (reference) | 0.000 | 0.913 | ||
| CG | 211 (47.4) | 48 (26.1) | 0.36 (0.21∼0.61) | ||||
| GG | 48 (10.8) | 45 (24.5) | 1.88 (1.00∼3.54) | ||||
| CG/GG vs. CC | 259 (58.2) | 93 (50.5) | 0.63 (0.40∼1.00) | 0.048 | 0.970 | ||
| CC/CG vs. GG | 397 (89.2) | 139 (75.5) | 2.09 (1.60∼5.26) | 0.000 | 0.970 | ||
| G allele freq. | 0.338 | 0.368 | 1.14 (0.90∼1.44) | 0.266 | 0.768 |
Table 4.
Genotype frequencies of the ADAM33 polymorphisms in dominant phenotypic model for patients with SLE




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download