Abstract
A fusion gene between echinoderm microtubule-associated protein-like 4 (EML4) and the anaplastic lymphoma kinase (ALK) has been identified in non-small cell lung cancers (NSCLCs). Although a few studies have evaluated EML4-ALK fusion genes in Korean NSCLCs, the prevalence of different EML4-ALK fusion variants has yet to be clearly assessed. Herein, we have examined the profiles of EML4-ALK fusion gene variants in Korean patients of NSCLCs. EML4-ALK fusion genes have been detected in 10 (6.0%) of 167 patients of NSCLCs and in 9 (7.4%) of 121 patients of adenocarcinoma. Of the 10 patients with fusion genes identified, 8 (80%) were E13;A20 (variant 1) and 2 (20%) were E6;A20, with an additional 33-bp sequence derived from intron 6 of EML4 (variant 3b). These results indicate that the profiles of EML4-ALK fusion gene variants in Korean patients of NSCLC may differ from those in other ethnic populations. Herein, we describe for the first time the profiles of EML4-ALK fusion variants of Korean patients with NSCLCs.
Non-small cell lung cancer (NSCLC) accounts for approximately 80%-85% of all cases of lung cancer, and is the leading cause of cancer deaths worldwide including Korea, with only 15% of patients surviving for more than 5 yr (1-3). Although cytotoxic chemotherapy remains the mainstay treatment for the majority of patients with advanced NSCLC, molecular-targeted therapy played an increasingly important role, particularly in genetically defined subsets of patients (4). Therefore, the identification of patients that harbor genetic alterations of the key oncogene for NSCLC is extremely important for selecting those most likely to derive benefit from a specific molecular targeted agent (5).
The echinoderm microtubule-associated protein-like 4 (EML4)-anaplastic lymphoma kinase (ALK) fusion oncogene resulting from the chromosome inversion inv(2) (p21; p23) represents a novel molecular target in NSCLC. Since its first report by Soda et al. (6), the EML4-ALK fusion gene has been indentified in 3%-7% of NSCLCs (7). Additionally, ALK kinase inhibitors have been developed and have been shown to be highly effective in NSCLCs harboring EML4-ALK fusion gene (8-10). Therefore, it is clearly imperative to identify NSCLC patients with EML4-ALK fusion gene (11).
Thus far, multiple EML4-ALK variants have been identified in NSCLCs (7, 12). All involve the intracellular tyrosine kinase domain of ALK beginning at the portion encoded for by exon 20. However, EML4 is variably truncated (occurring at exons 2, 6, 13, 14, 15, 18, and 20) and gives rise to diverse variants of EML4-ALK. Among the variants known thus far, it has been reported that E13;A20 (the nomenclature refers to the exon in EML4 [E] fused to the exon of ALK [A], variant 1) and E6a/b;A20 (variant 3a/b) are the most common variants, accounting for 33% and 29%, respectively, of all the EML4-ALK variants identified in NSCLCs (7, 12). Although a few studies have evaluated EML4-ALK fusion genes in Korean NSCLCs (13, 14), the prevalence of different EML4-ALK fusion variants has not been particularly well studied. Considering the differences in genetic and environmental factors related to lung cancer, it is possible that the profile of EML4-ALK fusion variants in the context of Korean lung cancer may differ from those of other countries. To answer this question, we evaluated EML4-ALK fusion variants using reverse-transcriptase-polymerase chain reaction (RT-PCR) in Korean NSCLCs.
Tumor and corresponding non-malignant lung tissue specimens were provided by the National Biobank of Korea - Kyungpook National University Hospital, Daegu, Korea, which is supported by the Ministry of Health, Welfare, and Family Affairs. All materials derived from the National Biobank were obtained in accordance with institutional review board-approved protocol, Kyungpook National University Medical Center (Approval No., KNUHBIO_10_1016).
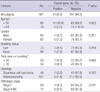
This study included 167 NSCLC patients who underwent curative resection at the Kyungpook National University Hospital, in Daegu, Korea between January 2001 and December 2009. Patients who underwent chemotherapy or radiotherapy prior to surgery were excluded in order to avoid effects on RNA. All patients included in this study were ethnic Koreans. This study included 46 patients with squamous cell carcinoma (SCC) and 121 patients with adenocarcinoma (AC). The study cohort comprised 85 males and 82 females. The patient population comprised 94 never-smokers and 73 smokers. Of the 121 patients with AC, 93 were never-smokers. All of the tumor and macroscopically-normal lung tissue samples were obtained at the time of surgery, then rapidly frozen in liquid nitrogen and stored at -80℃. Only tumors with greater than 80% of the tumor component were sent for analysis.
Total RNA was extracted from fresh frozen tissues using the RNeasy Mini kit (Qiagen Valencia, CA, USA), and the RNA extract was incubated with RNase-free DNase I (Qiagen) to remove contaminating DNA. Reverse transcription of total RNA was carried out using a Qiagen kit to generate complementary DNA (cDNA). In order to identify all possible EML4-ALK fusion cDNA, we conducted RT-PCR assays using two sense primers (5'-TCACTGTGCTAAAGGCGGCTTTGG-3', on exon 2 of EML4, and 5'-CCACACCTGGGAAAGGACCTAAAG-3', on exon 13 of EML4) and a single antisense primer (5'-CAGGGCTTCCATGAGGAAATCCAG-3', on exon 22 of ALK). PCR reactions were performed in a total volume of 20 µL containing 50 ng of cDNA, 0.2 mM of each primer, 0.2 mM dNTPs, 1 unit of Taq polymerase (Takara, Shuzo Co., Otus, Shiga, Japan), and 1 × reaction buffer (10 mM Tris-HCl [pH8.3], 50 mM KCl, and 1.5 mM MgCl2). The PCR cycle conditions consisted of an initial denaturation step at 95℃ for 5 min, followed by 35 cycles of 30 sec at 95℃; 30 sec at 66℃; 30 sec at 72℃; and a final elongation at 72℃ for 10 min. We used the primers 5'-GTCAGTGGTGGACCTGACCT-3' (forward) and 5'-TGAGCTTGACAAAGTGGTCT-3'(reverse) to amplify the glycer-aldehyde-3-phospate dehydrogenase (GAPDH) gene as an internal control. GAPDH amplification was carried out by preincubation for 5 min at 95℃ for initial denaturation, followed by 35 cycles of 30 sec at 95℃; 30 sec at 58℃; 30 sec at 72℃; and a final elongation at 72℃ for 10min. The PCR products were purified using a GENECLEAN Turbo kit (Q-Biogene, Carlsbad, CA, USA). Sequencing was performed using an ABI Prism 3100 Genetic Analyzer (PE Biosystems, Foster City, CA, USA). We also analyzed mutations in the EGFR (exons 18-21), ERBB2 (exons 19-20) and KRAS (exon 2) genes in the tumors harboring EML4-ALK fusion genes using PCR and direct sequencing, as described in our previous study (15).
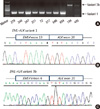
Using RT-PCR, EML4-ALK fusion transcripts were detected in 10 (6.0%) of the 167 NSCLCs. When the patients were stratified by median age, the fusion transcripts were significantly more common in younger patients than in older patients (10.8% vs 89.2%, P = 0.002). The fusion transcripts were more common in females, never-smokers, and ACs than in males, smokers, and SCCs, respectively, although not statistically significant (Table 1). All of 10 patients who harbored the EML4-ALK fusion transcripts had no mutations in EGFR, ERBB2, and KRAS genes. Nucleotide sequencing of the PCR products for the 10 identified positive cases revealed that 8 specimens (80%) harbored variant 1 (E13; A20), and 2 specimens (20%) harbored variant 3b (E6;A20 with an additional 33-bp sequence derived from intron 6 of EML4) (Fig. 1).
In this study, the frequency of EML4-ALK fusion genes in our series of NSCLCs was 6.0% (7.4% in ACs), which was consistent with the frequency of 3%-13% reported in East Asian patients with NSCLC (7, 12). Additionally, this was also similar to the frequency reported in a previous Korean study (4.2% of NSCLCs and 6.8% in ACs) (13), in which EML4-ALK fusion genes were analyzed by fluorescence in situ hybridization (FISH).
FISH is a standard method for detection of ALK rearrangement. However, unlike PCR, FISH cannot distinguish between different EML4-ALK fusion variants. Therefore, we employed RT-PCR assays to evaluate the profile of EML4-ALK fusion gene variants in Korean NSCLCs. Notably, the E13;A20 (variant 1) and E6a;A20 (variant 3b) account for 80% and 20%, respectively, of all the EML4-ALK variants identified in the current study. This frequency distribution of EML4-ALK fusion variants differed from those reported among other ethnic populations (7). Although all known variants have been demonstrated to possess potent oncogenic activity, it is possible that these different fusion variants have functional or therapeutic differences. Therefore, future studies will be required to clarify whether there are any functional or therapeutic differences amongst the different fusion variants.
One must consider a number of limitations of the present study. Because this study included only cases with available RNA, the demographic and clinicopathologic characteristics of the study population were somewhat different from those of a nationwide lung cancer survey (16). In addition, the results of RT-PCR analysis were not confirmed by FISH. Therefore, there might be false positive results (7).
Figures and Tables
References
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005. 55:74–108.
2. Jung KW, Park S, Kong HJ, Won YJ, Boo YK, Shin HR, Park EC, Lee JS. Cancer statistics in Korea: incidence, mortality and survival in 2006-2007. J Korean Med Sci. 2010. 25:1113–1121.
3. Jung KW, Won YJ, Park S, Kong HJ, Sung J, Shin HR, Park EC, Lee JS. Cancer statistics in Korea: incidence, mortality and survival in 2005. J Korean Med Sci. 2009. 24:995–1003.
4. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004. 350:2129–2139.
5. Dempke WC, Suto T, Reck M. Targeted therapies for non-small cell lung cancer. Lung Cancer. 2010. 67:257–274.
6. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small cell lung cancer. Nature. 2007. 448:561–566.
7. Sasaki T, Rodig SJ, Chirieac LR, Janne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010. 46:1773–1780.
8. Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas R, Lee J, Richards WG, Sugarbaker DJ, Ducko C, Lindeman N, Marcoux JP, Engelman JA, Gray NS, Lee C, Meyerson M, Jänne PA. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008. 14:4275–4283.
9. McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, Zhou W, Choi HG, Smith SL, Dowell L, Ulkus LE, Kuhlmann G, Greninger P, Christensen JG, Haber DA, Settleman J. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008. 68:3389–3395.
10. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Jänne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW, Iafrate AJ. Anaplastic lymphoma kinase inhibition in non-small cell lung cancer. N Engl J Med. 2010. 363:1693–1703.
11. Bronte G, Rizzo S, La Paglia L, Adamo V, Siragusa S, Ficorella C, Santini D, Bazan V, Colucci G, Gebbia N, Russo A. Driver mutations and differential sensitivity to targeted therapies: a new approach to the treatment of lung adenocarcinoma. Cancer Treat Rev. 2010. 36:S21–S29.
12. Horn L, Pao W. EML4-ALK: honing in on a new target in non-small cell lung cancer. J Clin Oncol. 2009. 27:4232–4235.
13. Kim H, Yoo SB, Choe JY, Paik JH, Xu X, Nitta H, Zhang W, Grogan TM, Lee CT, Jheon S, Chung JH. Detection of ALK gene rearrangement in non-small cell lung cancer: a comparison of fluorescence in situ hybridization and chromogenic in situ hybridization with correlation of ALK protein expression. J Thorac Oncol. 2011. 6:1359–1366.
14. Kim HR, Shim HS, Chung JH, Lee YJ, Hong YK, Rha SY, Kim SH, Ha SJ, Kim SK, Chung KY, Soo R, Kim JH, Cho BC. Distinct clinical features and outcomes in never-smokers with non-small cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer. 2011. Doi: 10.1002/cncr.26311.
15. Lee SY, Kim MJ, Jin G, Yoo SS, Park JY, Choi JE, Jeon HS, Cho S, Lee EB, Cha SI, Park TI, Kim CH, Jung TH, Park JY. Somatic mutations in epidermal growth factor receptor signaling pathway genes in non-small cell lung cancers. J Thorac Oncol. 2010. 5:1734–1740.
16. Lee C, Kang KH, Koh Y, Chang J, Chung HS, Park SK, Yoo K, Song JS. Characteristics of lung cancer in Korea, 1997. Lung Cancer. 2000. 30:15–22.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download