Abstract
Genetic information such as DNA sequences has been limited to fully explain mechanisms of gene regulation and disease process. Epigenetic mechanisms, which include DNA methylation, histone modification and non-coding RNAs, can regulate gene expression and affect progression of disease. Although studies focused on epigenetics are being actively investigated in the field of medicine and biology, epigenetics in dental research is at the early stages. However, studies on epigenetics in dentistry deserve attention because epigenetic mechanisms play important roles in gene expression during tooth development and may affect oral diseases. In addition, understanding of epigenetic alteration is important for developing new therapeutic methods. This review article aims to outline the general features of epigenetic mechanisms and describe its future implications in the field of dentistry.
The determination of the structure and function of deoxyribonucleic acid (DNA) opened up a new era in the fields of genomics and biotechnology. Geneticists can now precisely identify and determine the position of the specific gene within the genome that causes a certain genetic disease, thereby opening the door for potential cure for several diseases. However, the basic structure and function of DNA does not completely explain all of the underlying mechanisms of gene regulation and disease development. The field of epigenetics is now taking center stage in pursuit of better understanding the genome and ultimately gene expression.
Epigenetics is the study of alterations in gene regulation not caused by changes in the DNA sequence; the genome can have functionally relevant modifications that do not change the nucleotide sequence. There are numerous definitions for the term 'epigenetics'. Historically, epigenetics was used to describe cases that could not be explained by genetic principles.1 Conrad Waddington, who coined the term, defined epigenetics as 'the branch of biology that studies the causal interaction between genes and their product, which bring the phenotype into being'.1 He suggested that the development of undifferentiated embryos could be changed by epigenetics.2 Holliday described epigenetics as 'the study of the mechanisms of temporal and spatial control of gene activity during the development of a complex organism'.3 According to Russo et al., epigenetics was defined as 'the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in the DNA sequence'.4 The Greek prefix 'epi-' in epigenetics means 'on the top of' or 'in addition to' genetics. In 2008, a consensual definition of epigenetics was established as a 'stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence'.5
The mechanisms underlying epigenetic alterations are complex and involve DNA methylation, histone modification, and gene regulation by non-coding RNAs.1,2,6,7,8 In addition, epigenetic modifications are potentially reversible and transient. Many environmental factors affect these mechanisms.9 Ultimately, epigenetic alteration modulates gene expression and affects various gene functions (Figure 1).
Studies related to epigenetic characteristics are being extensively researched in the field of medicine and biology, thereby being reported the effects of epigenetics on pathogenesis.6,7,8,10 However, epigenetics in dental research is at the early stages. Nevertheless, studies concerning the epigenetics in dentistry are worthy of notice because epigenetic mechanisms play important roles in gene expression during development and pathological processes of oral diseases.11 For example, epigenetic modifications may lead to dental abnormality during tooth development stage.12,13 Also, various epigenetic factors may induce dental differences in monozygotic twin pairs.14,15 Many studies have reported that inflammatory reactions in infected pulp and periodontal tissue affect epigenetic modifications causing change of gene expression.16,17,18,19,20,21,22,23,24 Recently, changes of epigenetic biomarkers, such as inflammatory cytokines, involved in periodontitis are being reported.17,18,19,20,21,22 Furthermore, epigenetic mechanisms related to pulp repair and regeneration are being studied.25,26,27
Understanding epigenetic changes may be useful for developing new therapeutic methods to treat genetic diseases, and thus they may have multiple applications in the fields of dentistry. Therefore, this review article aims to outline the general features of epigenetic mechanisms, and describe its future implications in the field of dentistry.
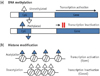
DNA methylation is the most characterized type of chromatin modification. The process of DNA methylation involves the covalent transfer of a methyl group from S-adenosyl methionine (SAM) to cytosines present in cytosine-phosphate-guanine (CpG) dinucleotides of the DNA chain.6 CpG sequences located throughout the eukaryotic genome are usually highly methylated and associated with repetitive sequences. These sequences tend to be germline-specific or associated with imprinted genes, and the CpG methylation results in transcriptional repression of the surrounding transcripts (Figure 2a).28
Mammalian genomes are composed of a large number of CpG sites, termed as the CpG islands.6 CpG islands usually lack DNA methylation and are related to the gene promoters region.6 DNA methyltransferases (DNMTs) establish and maintain DNA methylation patterns using SAM as the methyl donor.7 These methyl groups from the resulting methylated cytosine stick out into the major groove of DNA and serve to block the binding of the transcription factors to DNA.29 Additionally, the exposed methylation sites allow for interaction with methyl-binding proteins, such as the methyl-CpG-binding domain proteins (MBDs).30 These proteins influence chromatin condensation by assembling histone deacetylases. Histone deacetylases remove the acetyl group from histones, allowing for the histones to wrap the DNA more tightly.31 This process leads to gene silencing and chromatin compaction.32 DNA methylation patterns affect gene expression strongly, and these epigenetic patterns are conserved through cellular progeny by DNA replication.7 Abnormal methylation states can therefore lead to disease development. For instance, recent studies have shown that hypomethylation changes in DNA are associated with chromosome instability and activation of transposable elements in human cancers.33 Thus, DNA methylation is critical for proper regulation of the genome. According to recent studies in the field of dentistry, DNA methylation patterns can be altered by a persistent inflammation.16,17,22 Changes of DNA methylation patterns and cytokine gene expression can be observed in chronic periodontitis.17,18,19,20 In addition, methylation patterns may be different between healthy and inflamed dental pulp.23
The nucleosome, which is the basic unit of chromatin, consists of a DNA segment and eight core histones. This histone octamer consists two copies of each of H2A, H2B, H3, and H4. This composition provides a rigid structure to chromatin. Through post-translational modification of the core histones, chromatin is either condensed or relaxed, thus regulating gene transcription and serving as a crucial epigenetic mechanism.
The modification of histones takes place mostly at the N-terminal tails of the protein.34 Acetylation of the core histones results in an 'open' chromatin conformation that facilitates transcription (Figure 2b).35 The acetylated N-termini, which protrude from the nucleosome core, promote a more relaxed structure of the chromatin, allowing for the recruitment of the basic transcription factors.6 Conversely, histone deacetylases remove the acetyl groups, causing the chromatin to become more condensed and repress gene transcription (Figure 2b).6 On the other hand, histone methylation can either result in an activated or a repressed chromatin state.6 For example, tri-methylation of histone H3 on lysine residues 4 and 36 promotes an open chromatin structure, leading to active transcription.36 However, histone methylation on lysine residues 9 and 27 leads to condensation of the chromatin and thus, gene silencing.37 In summary, histone post-translational modification is a powerful epigenetic mechanism since it can regulate gene expression by altering chromatin states through either acetylation or methylation.
Recent studies have reported that histone modifications may induce differentiation and mineralization in dental pulp stem cells.25,26,38 Histone acetylations and deacetylations play crucial roles in regulations of gene expression and may promote pulp repair and regeneration.25,26,38,39,40 Therefore, studies on epigenetic regulations in restorative dentistry have grown in importance.
Non-coding RNAs are functionally relevant RNA molecules, despite not encoding for a protein. This group of RNAs includes a wide range of important RNAs such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), microRNAs (miRNAs), and short-interfering RNAs (siRNAs). The latter RNAs, miRNAs and siRNAs, have been shown to regulate gene expression without altering the DNA sequence. For instance, miRNAs are small single-strand non-coding RNAs (20 - 24 nucleotides) that negatively regulate the expression of their target genes at the post-transcriptional level.6,8 MiRNAs binds to the 3' untranslated region (3'UTR) of their target messenger RNAs (mRNAs), and lead to subsequent degradation or translational repression of the bound mRNA through recruitment of the RNA-induced silencing complex.6 Guo et al. suggested that reduction of the protein level by endogenous miRNAs is caused by destabilization of the target mRNA.41 Furthermore, recent studies have reported that miRNAs are involved in multiple vital processes, throughout the regulation of development or differentiation of a disease.6,8,41 Studies in dentistry have reported that non-coding RNAs are involved in oral diseases such as specific syndromes, oral cancer and oral immunology.42,43 In addition, recent studies have demonstrated that miRNAs play essential roles in odontoblast differentiation.42,43
Researchers are now actively studying epigenetic changes seen during the initiation, development, and metastatic stages of cancer, in attempts to develop better diagnostic tools and therapies for patients. Since 2008, epigenomics has been one of the most ambitious projects at the US National Institutes of Health (NIH). It has been reported that there are epigenetic changes during fetal development, progression of cancer states, or in chronic diseases such as autoimmune diseases, diabetes mellitus, cardiovascular diseases, and mental diseases in adults.10 The following sections will discuss the representative epigenetic mechanisms related to gene regulation.
Diploid organisms receive two copies of each gene, one from each parent. Mostly, both copies are either repressed or transcribed identically. Investigators have suggested that genes inherited by each parent have been marked or imprinted permanently.7 Therefore, patterns of expression, depending on the maternal and paternal inheritance, will exhibit a mosaic pattern from the parents.7 In mammals, genomic imprinting reflects that expression of alleles at certain gene loci is not equivalent but is determined by the parent of origin.44 For example, researchers identified that insulin-like growth factor-2 receptor (IGF2R) and H19 are active only when inherited from the mother. On the other hand, insulin-like growth factor-2 (IGF2) is expressed only when passed down from the father.7
DNA methylation is considered to be the main mechanism underlying imprinting. During this process, one copy of a gene is marked with DNA methylation depending on parental origin, and this methylation is maintained during cell division by 5-cytosine DNA methytransferase-1 (DNMT1).45,46 DNMT1 carries out methylation in hemimethylated CpG areas and these methylated patterns are replicated to the newly synthesized DNA strands.7 IGF2 imprinting is regulated during fetal development and is a good representative example of the process of imprinting.7 IGF2 is an essential somatic growth factor for the fetus, and any damage or misregulation could have detrimental consequences. Therefore, the epigenetic program which regulates IGF2 gene expression is an important component for proper development.
Up to now, studies on dental diseases related to imprinting are rare. However, imprinting can be connected with dental diseases such as developmental anomalies and defects because DNA methylation is the key mechanism on both sides.11
Somatic epigenetic inheritance, like DNA methylation and chromatin remodeling patterns, is very crucial in the development of multicellular eukaryotic organisms. Although the gene sequence is static, cells differentiate into many different types. They carry out dissimilar functions and respond divergently to the environment and intercellular signaling. Therefore, the underlying epigenetic processes are the key to the various types of cellular differentiation and function.
Recent studies have reported that certain cell lineages regulate the gene expression pattern sequentially by an epigenetic program.47,48 For example, the T-helper cell population of the immune system is regulated by this epigenetic programming.48 When cluster of differentiation (CD4+) T-cells are mature, interferon gamma (IFNγ) gene is epigenetically activated and the Interleukin 4 (IL4) gene is epigenetically silenced.7,48 This mechanism leads to a progressive polarization of T-cell responses, as epigenetic modifications are modified by antigenic and cytokine actions. Therefore, different T-helper cells are made, and they maintain a polarized phenotype.7
Epigenetic mechanisms also play a crucial role in tooth development.13 For instance, histone demethylase may regulate the dental stem cell differentiation.13 In addition, histone acetyltransferase and non-coding RNAs may influence odontogenic differentiation.40,42 Epigenetic factors present at each developmental stage can affect the developmental processes.13 Thus, some studies have proposed that epigenetic events during tooth development may lead to dental differences in monozygotic twins having identical genotypes.14,15
Modification of DNA methylation has been shown to begin as early as the prenatal stage due to environmental factors. For example, the methylation of fetal DNA can be altered due to low dietary levels of folate, methionine, or selenium in utero, and this can persist well into adulthood.49,50,51 According to Baker et al., intrauterine exposures can cause fetal programming that lasts even in adulthood, and may increase the risk of adult diseases such as cardiovascular disease and type 2 diabetes.52 Therefore, intrauterine nutrition can have a significant effect on the epigenetic programming of the fetus. For example, methyltetrahydrofolate is an essential methyl donor for SAM which is used by the enzyme, DNMT, to methylate CpG residues.53 During pregnancy, maternal folate deficiency causes inadequate levels of SAM.45 Consequentially, maternal folate deficiency can lead to DNA hypomethylation, which can cause excessive expression of certain genes and genetic instability in the fetus.51 Additionally, multiple dietary or other environmental factors can influence epigenetic modifications throughout life. Recent studies have reported that environmental factors may increase the attachment loss and alter the virulence of pathogens in periodontal tissues.54,55 Furthermore, epigenetic alteration may affect oral health and inflammatory conditions.9
Inflammation is a biological response to noxious stimuli such as pathogens or irritants. Many studies suggest inflammation as a cause of epigenetic changes, including DNA methylation, histone modification, and targeting by microRNAs.6,7,9,16,17,18 Some studies have shown that the activation of immune responses will lead to potential epigenetic changes.16,17,18,56 Ito reported that inflammatory signals promote the activity of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), thereby potentially modifying histone methylation patterns and promoting gene expression.56 These data have potential implications for diseases such as periodontitis, a typical chronic inflammatory disease. Several studies have reported that infected pulp and periodontal tissue may alter gene expression patterns of inflammatory cytokines.17,18,19,20,21,22,23 Therefore, roles of these alterations as epigenetic biomarkers are necessary to be determined for prevention and treatment of dental diseases.
DNA methylation is a well-characterized epigenetic modification seen in cancer. These epigenetic changes are verified as key factors of carcinogenesis. In most tumors, hypomethylation occurs, which increases transcriptional activity. This often occurs at an unstable sequence and is related to increased tumor frequency. It has been regarded as the earliest epigenetic modification signifying changes from normal to pre-malignant cells.7 In contrast, some studies have reported that hypermethylation of tumor suppressor gene is also related to carcinogenesis.57,58 Hypermethylation of promoters for tumor-suppressor genes leads to gene repression and subsequent tumor progression.59 It has been suggested that oncogenesis may originally stem from epigenetic disturbance. In a recent review, Choi and Myers reported that genetic and epigenetic modifications may be the basis for the molecular pathogenesis of oral squamous cell carcinoma and the role of tumor suppressor genes such as p53, p16 and p21 in cancer.60
Recently, many studies have examined epigenetics in the context of dental health.9,12,13,14,16,17,18,20,25 Lod et al. described epigenetic alterations associated with oral health and inflammatory conditions.9 Brook proposed that dental anomalies are results of genetic-epigenetic interactions.13 Several nutritional factors such as folate, vitamin B12, and vitamin A may lead to changes in epigenetic modification. Additionally, smoking also causes hypomethylation and hypermethylation changes in DNA.9 Therefore, some exogenous factors such as diet, smoking, environment, bacteria, inflammation, and age may affect oral health by causing epigenetic changes (Figure 3). Some studies have shown that epigenetic changes may affect dental differences in monozygotic twin pairs.14,15 These studies indicate that minor epigenetic variations during odontogenesis may lead to distinct differences.14,15 For example, Fan et al. found that the oculofaciocardiodental (OFCD) syndrome, which is characterized by canine teeth with extremely long roots, is associated with a BCL-6 corepressor (BCOR) mutation.12 This mutation leads to the upregulation of AP-2a in mesenchymal stem cells and promotes osteodentinogenesis. Fan et al. claimed that BCOR plays an important role in the development and maintenance of homeostasis through histone methylation.12
The next sections cover the epigenetic studies in periodontal disease and dental pulp cells. These fields are the subjects of significant ongoing research.
Periodontal disease is a complex infection characterized by inflammation and destruction of the tooth-supporting tissue. Recently, a review article has highlighted that gene expression was altered by epigenetic modifications in periodontitis.17 Gomez et al. reported that the methylation pattern caused by changes in cytokine gene expression could lead to inflammatory diseases.16 Inflammatory cytokines such as IL1, IL4, IL6, and IL10 are found to be overexpressed in the inflamed periodontal system.21 Stenvinkel et al. suggested that a persistent inflammation leads to DNA methylation, silencing the suppressors of cytokine signaling and inducing the active expression of cytokine signaling.22 Cytokines such as IL6 and IFNγ were found to be overexpressed in inflamed tissues of chronic periodontitis patients.18,61 Zhang et al. reported that hypomethylation of the IFNγ promoter may lead to increased IFNγ transcription in chronic periodontitis, thus resulting in IFNγ overexpression.18 In contrast, genes like tumor necrosis factor alpha (TNFa) and cyclooxygenase-2 (COX-2) were hypermethylated at the CpG site which represses expression.17 Zhang et al. reported that the change in the methylation patterns may be a crucial factor in regulating TNFa transcription in periodontitis.19 Additionally, it was found that chronic inflammation is related to altered DNA methylation levels and may lead to a decreased expression of COX-2.20 Taken together, the identification of genetic factors and epigenetic factors could be valuable for developing oral treatments and for disease prevention.17
The regeneration of pulp is very important in the treatment process of damaged pulp cells. In restorative dentistry, tissue regeneration has become increasingly important due to changing treatment concepts. It has been attempted gradually in clinical applications.
Recently, Duncan et al. reported that epigenetic modification may affect dental pulp cell behavior.25,26 These studies suggested that histone deacetylase inhibitors (HDACis) epigenetically promote reparative events in dental pulp cells by reducing proliferation and increasing mineralization.25,26 These studies reported that HDACis stimulate osteopontin and bone morphogenic protein 2 (BMP 2), thereby promoting mineralization and differentiation events.25,26 Paino et al. suggested that HDACis lead to promote expression of osteopontin and bone sialoprotein but downregulate expression of osteocalcin.39 In addition, Wang et al. suggested that histone acetyltransferase p300 modulates expression of odontogenic marker genes, such as dentin matrix protein 1, dentin sialophosphoprotein, dentin sialoprotein, osteopontin and osteocalcin, and modifies odontogenic differentiation in dental pulp stem cells.40 Therefore, epigenetic alterations may play important roles in regulating the expression of core genes in dental pulp cells.
HDACis are known to regulate the homeostatic balance by modulating transcription and cell behavior.62,63 Representatively, they modulate the balance between histone acetyltransferase and histone deacetylases, which lead to gene expression change.25,26,38 Because of their characteristics, HDACis may be used as clinical tools in the treatment of cancer, chronic inflammation and stem cell engineering.38 Thus, they can be applied in clinical practices including inflammation control, mineralization induction, repair and regeneration in the field of restorative dentistry. Duncan et al. suggested that HDACis have the potential for use of therapeutic materials in dental restorative treatments.38 Recently, Hui et al. proposed that enhancer of zeste homolog 2 (EZH2) may work as a regulator in dental pulp inflammation, proliferation and regeneration.27 EZH2 involved in histone modification mechanism may act as epigenetic markers.27 This study suggested that EZH2 can be applied to dental pulp regeneration through further studies about precise mechanisms.27
Cardoso et al. investigated the changes in the methylation patterns in human dental pulp cells.23,24 They found that the methylation patterns of IFNγ was different between healthy and inflamed human pulp, where IFNγ methylation is reduced in the inflamed pulp.23 Recently, Cardoso et al. compared the DNA methylation patterns for two other genes, namely, Toll-like receptors 2 (TLR2) and CD14, between healthy and inlfamed human dental pulp.24 However, no significant differences were found between the two groups.24 Sun et al. reviewed that non-coding RNAs may affect odontoblast differentiation and oral diseases.42 It proposes that epigenetic mechanisms may cause dental anomalies and defects such as dentin dysplasia and dentinogenesis imperfecta.42 Furthermore, dental genetic defects such as cleft lip, cleft palate and syndromes may be related to epigenetic modification during odontoblast differentiation in dental stem cells.11 Therefore, further studies are needed to determine roles of epigenetic factors in dental anomalies and to apply clinically.
This review presents evidence that epigenetics plays an important role in gene regulation. Relevant epigenetic mechanisms include DNA methylation, histone modification, and non-coding RNAs. The roles of these mechanisms are varied but they ultimately affect gene expression. Epigenetic modification can cause gene imprinting and regulate development in eukaryotic organisms. Furthermore, exogenous factors such as diet, smoking, stimuli, and inflammation can cause alterations in epigenetically regulated gene expression. Epigenetic changes can contribute to the progression of certain diseases such as cancer.
Though studies focused on epigenetics in dentistry are still in the early stages, there is increasingly more evidence for the association between epigenetic changes and periodontal diseases, as well as inflamed dental pulp cells. Future researches are required to corroborate these initial studies through which better understanding of the effect of epigenetics on dental health, and development of effective therapies may be established.
Figures and Tables
Figure 1
Environmental and epigenetic factors. Many environmental factors such as diet, smoking, inflammation, stimuli and age may affect gene regulation, which leads to epigenetic modification in the genome. The mechanisms underlying epigenetic modification involve DNA methylation, histone modification, and gene regulation by non-coding RNAs. These mechanisms modulate gene expression and affect various gene functions.
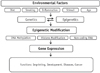
Figure 2
Epigenetic mechanisms. (a) DNA methylation, Methylated DNA sequences in CpG sites cause the more condensed DNA structure. This process leads to transcriptional repression and gene silencing; (b) Histone modification, Acetylation of histones results in an open chromatin conformation, allowing for the recruitment of the basic transcription factors and this process facilitates gene transcription. In contrast, histone deacetylases remove the acetyl groups, causing the chromatin to become more condensed, and they repress gene transcription. CpG, cytosine-phosphate-guanine.
Acknowledgement
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2012-009268).
References
1. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007; 128:635–638.

3. Holliday R. Mechanisms for the control of gene activity during development. Biol Rev Camb Philos Soc. 1990; 65:431–471.

4. Russo VE, Martienssen RA, Riggs AD. Epigenetic mechanisms of gene regulation. New York: Cold Spring Harbor Laboratory Press;1996. p. 1–4.
5. Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009; 23:781–783.
7. Barros SP, Offenbacher S. Epigenetics: connecting environment and genotype to phenotype and disease. J Dent Res. 2009; 88:400–408.

8. Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011; 90:430–440.

9. Lod S, Johansson T, Abrahamsson KH, Larsson L. The influence of epigenetics in relation to oral health. Int J Dent Hyg. 2014; 12:48–54.

11. Williams SD, Hughes TE, Adler CJ, Brook AH, Townsend GC. Epigenetics: a new frontier in dentistry. Aust Dent J. 2014; 59:Suppl 1. 23–33.

12. Fan Z, Yamaza T, Lee JS, Yu J, Wang S, Fan G, Shi S, Wang CY. BCOR regulates mesenchymal stem cell function by epigenetic mechanisms. Nat Cell Biol. 2009; 11:1002–1009.

13. Brook AH. Multilevel complex interactions between genetic, epigenetic and environmental factors in the aetiology of anomalies of dental development. Arch Oral Biol. 2009; 54:Supplement 1. S3–S17.

14. Townsend GC, Richards L, Hughes T, Pinkerton S, Schwerdt W. Epigenetic influences may explain dental differences in monozygotic twin pairs. Aust Dent J. 2005; 50:95–100.

15. Townsend G, Bockmann M, Hughes T, Brook A. Genetic, environmental and epigenetic influences on variation in human tooth number, size and shape. Odontology. 2012; 100:1–9.

16. Gomez RS, Dutra WO, Moreira PR. Epigenetics and periodontal disease: future perspectives. Inflamm Res. 2009; 58:625–629.

17. Lindroth AM, Park YJ. Epigenetic biomarkers: a step forward for understanding periodontitis. J Periodontal Implant Sci. 2013; 43:111–120.

18. Zhang S, Crivello A, Offenbacher S, Moretti A, Paquette DW, Barros SP. Interferon-gamma promoter hypomethylation and increased expression in chronic periodontitis. J Clin Periodontol. 2010; 37:953–961.

19. Zhang S, Barros SP, Moretti AJ, Yu N, Zhou J, Preisser JS, Niculescu MD, Offenbacher S. Epigenetic regulation of TNFA expression in periodontal disease. J Periodontol. 2013; 84:1606–1616.
20. Zhang S, Barros SP, Niculescu MD, Moretti AJ, Preisser JS, Offenbacher S. Alteration of PTGS2 promoter methylation in chronic periodontitis. J Dent Res. 2010; 89:133–137.

21. Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med. 2003; 14:430–449.

22. Stenvinkel P, Karimi M, Johansson S, Axelsson J, Suliman M, Lindholm B, Heimbürger O, Barany P, Alvestrand A, Nordfors L, Qureshi AR, Ekström TJ, Schalling M. Impact of inflammation on epigenetic DNA methylation - a novel risk factor for cardiovascular disease? J Intern Med. 2007; 261:488–499.

23. Cardoso FP, Viana MB, Sobrinho AP, Diniz MG, Brito JA, Gomes CC, Moreira PR, Gomez RS. Methylation pattern of the IFN-gamma gene in human dental pulp. J Endod. 2010; 36:642–646.
24. Cardoso FP, de Faria Amormino SA, Dutra WO, Ribeiro Sobrinho AP, Moreira PR. Methylation pattern of the CD14 and TLR2 genes in human dental pulp. J Endod. 2014; 40:384–386.
25. Duncan HF, Smith AJ, Fleming GJ, Cooper PR. Histone deacetylase inhibitors induced differentiation and accelerated mineralization of pulp-derived cells. J Endod. 2012; 38:339–345.

26. Duncan HF, Smith AJ, Fleming GJ, Cooper PR. Histone deacetylase inhibitors epigenetically promote reparative events in primary dental pulp cells. Exp Cell Res. 2013; 319:1534–1543.

27. Hui T, A P, Zhao Y, Wang C, Gao B, Zhang P, Wang J, Zhou X, Ye L. EZH2, a potential regulator of dental pulp inflammation and regeneration. J Endod. 2014; 40:1132–1138.

28. Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009; 10:805–811.

29. Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000; 405:486–489.
30. Loenen WA. S-adenosylmethionine: jack of all trades and master of everything? Biochem Soc Trans. 2006; 34:330–333.

31. Vucic EA, Brown CJ, Lam WL. Epigenetics of cancer progression. Pharmacogenomics. 2008; 9:215–234.

32. Bird AP, Wolffe AP. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999; 99:451–454.

33. Cheung HH, Lee TL, Rennert OM, Chan WY. DNA methylation of cancer genome. Birth Defects Res C Embryo Today. 2009; 87:335–350.

34. Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns-from conservation to diversity. Trends Plant Sci. 2006; 11:199–208.

36. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837.

37. Lan F, Shi Y. Epigenetic regulation: methylation of histone and non-histone proteins. Sci China C Life Sci. 2009; 52:311–322.

38. Duncan HF, Smith AJ, Fleming GJ, Cooper PR. HDACi: cellular effects, opportunities for restorative dentistry. J Dent Res. 2011; 90:1377–1388.
39. Paino F, La Noce M, Tirino V, Naddeo P, Desiderio V, Pirozzi G, De Rosa A, Laino L, Altucci L, Papaccio G. Histone deacetylase inhibition with valproic acid downregulates osteocalcin gene expression in human dental pulp stem cells and osteoblasts: evidence for HDAC2 involvement. Stem Cells. 2014; 32:279–289.

40. Wang T, Liu H, Ning Y, Xu Q. The histone acetyltransferase p300 regulates the expression of pluripotency factors and odontogenic differentiation of human dental pulp cells. PLoS One. 2014; 9:e102117.

41. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010; 466:835–840.

42. Sun Q, Liu H, Chen Z. The fine tuning role of microRNA-RNA interaction in odontoblast differentiation and disease. Oral Dis. 2014; 03. 22. doi: 10.1111/odi.12237. [Epub ahead of print].

43. Perez P, Jang SI, Alevizos I. Emerging landscape of non-coding RNAs in oral health and disease. Oral Dis. 2014; 20:226–235.

45. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999; 99:247–257.

46. Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007; 213:384–390.

47. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007; 447:425–432.

48. Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003; 4:616–623.

49. Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, Milliken EE, Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999; 43:985–991.

50. Lund G, Andersson L, Lauria M, Lindholm M, Fraga MF, Villar-Garea A, Ballestar E, Esteller M, Zaina S. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004; 279:29147–29154.

51. Zaina S, Lindholm MW, Lund G. Nutrition and aberrant DNA methylation patterns in atherosclerosis: more than just hyperhomocysteinemia? J Nutr. 2005; 135:5–8.

52. Barker DJ, Eriksson JG, Forsén T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol. 2002; 31:1235–1239.

54. Ohi T, Uehara Y, Takatsu M, Watanabe M, Ono T. Hypermethylation of CpGs in the promoter of the COL1A1 gene in the aged periodontal ligament. J Dent Res. 2006; 85:245–250.

55. Wu H, Lippmann JE, Oza JP, Zeng M, Fives-Taylor P, Reich NO. Inactivation of DNA adenine methyltransferase alters virulence factors in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2006; 21:238–244.

56. Ito K. Impact of post-translational modifications of proteins on the inflammatory process. Biochem Soc Trans. 2007; 35:281–283.

57. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003; 349:2042–2054.

59. Breivik J, Gaudernack G. Genomic instability, DNA methylation, and natural selection in colorectal carcinogenesis. Semin Cancer Biol. 1999; 9:245–254.

60. Choi S, Myers JN. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008; 87:14–32.

61. Babel N, Cherepnev G, Babel D, Tropmann A, Hammer M, Volk HD, Reinke P. Analysis of tumor necrosis factor-alpha, transforming growth factor-beta, interleukin-10, IL-6, and interferon-gamma gene polymorphisms in patients with chronic periodontitis. J Periodontol. 2006; 77:1978–1983.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download