Abstract
Background/Aims
The incidence and severity of Clostridium difficile infection (CDI) have increased worldwide, resulting in a need for rapid and accurate diagnostic methods.
Methods
A retrospective study was conducted to compare CDI diagnosis methods between January 2014 and December 2014. The stool samples, which were obtained in presumptive CDI patients, were compared for their diagnostic accuracy and rapidity, including real-time polymerase chain reaction (PCR) of toxin genes, C. difficile toxin assay, and culture for C. difficile.
Results
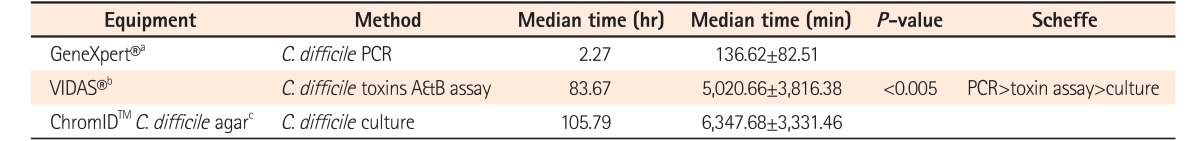
A total of 207 cases from 116 patients were enrolled in this study and 117 cases (56.5%) were diagnosed as having CDI. Among the 117 cases, the sensitivities of real-time PCR, C. difficile toxin assay, and culture for C. difficile were 87.2% (102 cases; 95% CI, 80.7%–92.8%), 48.7% (57 cases; 95% CI, 41.0%–59.8%), and 65.0% (76 cases; 95% CI, 60.2%–78.5%), respectively (P<0.005). Notably, 34 cases (29.0%) were diagnosed with CDI by real-time PCR only. The time required to obtain results was 2.27 hours (136.62±82.51 minutes) for real-time PCR, 83.67 hours (5,020.66±3,816.38 minutes) for toxin assay, and 105.79 hours (6,347.68±3,331.46 minutes) for culture (P<0.005), respectively.
As the number of long-term inpatients and the prevalence of chronic diseases increase, nosocomial infections with certain bacterial and viral infections are increasing worldwide. Clostridium difficile infection (CDI) is an infectious disease frequently encountered in clinical practice. Recently, the incidence and severity of CDI has increased markedly worldwide.1
2
3
4
5 Severe CDI associated with an emerging epidemic strain, PCR-ribotype 027, has exhibited an increasing frequency and rate of mortality.1
4 In addition, recent studies have reported high recurrence rates of CDI (19.5%) at 8 weeks after successful treatment.6 Therefore, effective diagnostic methods and treatment modalities are necessary.
CDI was suspected in recently hospitalized patients or inpatients with prior diarrhea, abdominal pain, or fever and those who underwent antibiotic therapy.7
8 The diagnostic methods of CDI include the stool toxin assay, culture for C. difficile, and PCR for identifying the C. difficile chromosomal genes tcdA or tcdB, which encode C. difficile toxin A or B, respectively. The recent report about diagnostic methods for early diagnosis was insufficient for comparison of the increasing CDI burden with the severity of the disease. Therefore, our study sought to compare the sensitivity and specificity of diagnostic methods and investigated the time required to obtain the results.
This study investigated CDI-related diagnostic tests by comparing the sensitivity of the methods that were performed on the stool samples obtained from CDI patients from January 1 to December 31, 2014 at Seoul Paik Hospital. For patients with CDI, baseline characteristics, including underlying diseases and drugs administered to patients, were reviewed retrospectively. The stool samples that were obtained in presumptive CDI patients were compared to determine diagnostic accuracy and rapidity, including real-time PCR of toxin genes, C. difficile toxin assay, and culture for C. difficile. When any one of these methods could not be performed, these cases were excluded. In addition, toxin assay, which showed equivocal results, were excluded. This study was approved by the Institutional Review Board of Seoul Paik Hospital (IRB No. IIT-2015-408). A consent waiver was granted for this database study.
Diarrhea was defined as a bowel habit change with more than 3 unformed stools per day for more than 2 days. CDI was defined as the presence of diarrhea and any positive outcome (1 or more) on the following diagnostic tests: (1) C. difficile toxin A and/or B detection by enzyme immunoassay, (2) positive stool culture for C. difficile, and (3) positive stool PCR for C. difficile toxin B.
Collected stool samples were immediately transported to the laboratory and stored at 2℃ to 8℃ prior to testing. The primer and probe of the GeneXpert® (Cepheid, Sunnyvale, CA, USA) C. difficile test system were used. This system targets the toxin B gene (tcdB), the critical component of all toxigenic C. difficile strains. Furthermore, it detects the epidemic 027/NAP1/BI strain. The GeneXpert® C. difficile test system automates and integrates sample purification, nucleic acid amplification, and detection of the target sequences in simple or complex samples using real-time PCR. The result is represented as toxigenic C. difficile positive or negative and 027/NAP1/BI presumptive positive or negative.
The stool samples were tested with VIDAS® C. difficile toxin assay A and/or B (bioMérieux, Marcy L'Etoile, France). If toxin A and/or B is present in the stool sample they would adhere to the biotin coated interior wall of the solid-phase receptacle, coupled with anti-toxin A antibody (rabbit polyclonal) and anti-toxin B antibody (mouse monoclonal). If the relative fluorescence value (RFV) is more than 0.37, the test is positive for CDI; an RFV of 0.13 to less than 0.37 means the result is equivocal, and if the RFV is less than 0.13, the test is negative.
The stool samples were inoculated into ChromID™ C. difficile agar (bioMérieux) consisting of nutrient-rich material with a combination of multiple peptone and taurocholate, which promotes spores germination. Then the agar was anaerobically incubated for 48 to 72 hours. The identification of suspected C. difficile was conducted through analysis of Gram staining, spore staining, and/or a biochemical assay using an ANA identification test kit (bioMérieux).
The statistical analysis was performed using PASW version 18.0 (SPSS Inc., Chicago, IL, USA). The outputs were relative positive fractions with 95% CIs. The accuracy of the diagnosis methods was determined using a McNemar's test. A P-value of 0.005 was considered significant. The time that was required to obtain results was analyzed by ANOVA. In addition, we used the Mann-Whitney test or chi-square test to compare the 2 groups.
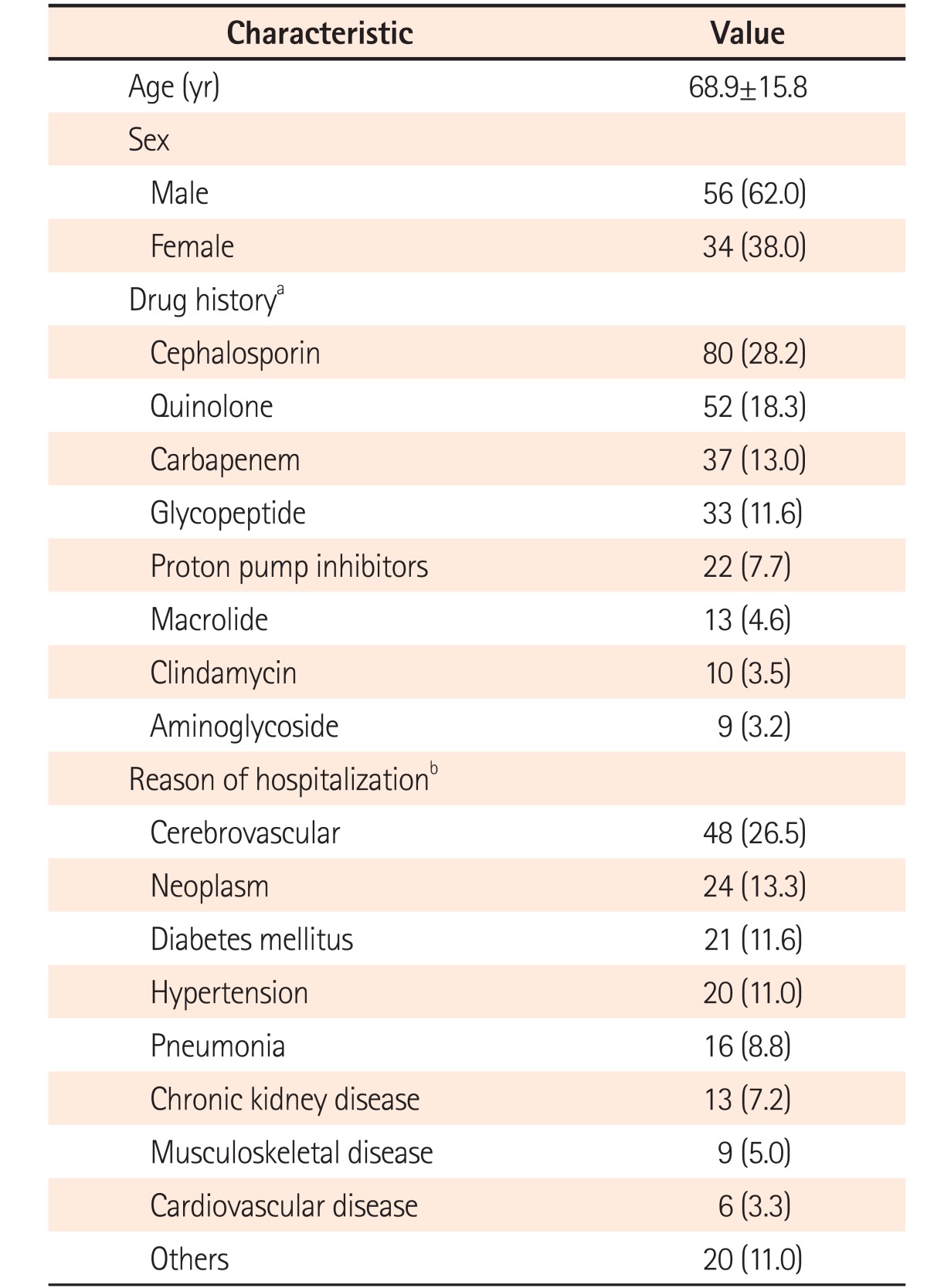
A total of 452 cases were initially enrolled in this study and 225 cases were excluded, because the 3 methods could not be performed simultaneously. In addition, 20 cases showed equivocal results of the toxin assay, leading to exclusion. Therefore, a total of 207 cases from 116 patients were investigated in this study and from these, 117 cases from 90 patients were diagnosed as CDI (Fig. 1). The patients comprised 56 men (62.0%) and 34 women (38.0%), with their mean age being 68.9±15.8 years (Table 1).
A total of 181 underlying diseases were counted in 90 patients, of which some had more than 1 underlying disease. The most common underlying disease among the patients with CDI was cerebrovascular disease (48 cases, 26.5%), while neoplastic disease was present in 24 cases (13.3%). Regarding antibiotics administration, some of the patients received more than 1. The most common antibiotic was cephalosporins (80 cases, 28.2%), followed by quinolones (52 case, 18.3%). Notably, 22 cases (7.7%) were related to proton pump inhibitors (Table 1).
The required time to obtain results was measured from the beginning of the test to when the results were obtained. The required time was 2.27 hours (136.62±82.51 minutes) for real-time PCR, 83.67 hours (5,020.66±3,816.38 minutes) for toxin A & B assay, and 105.79 hours (6,347.68±3,331.46 minutes) for C. difficile culture (P<0.005), respectively. The PCR analysis was the most rapid test compared to the other methods and showed a statistically significant difference (P<0.005) (Table 2). The rapidity of the real-time PCR method over the toxin assay and the culture method was confirmed using a Scheffe test.
Among the 117 cases, the sensitivities of real-time PCR, C. difficile toxin assay and culture for C. difficile were 87.2% (102 cases; 95% CI, 80.7%–92.8%), 48.7% (57 cases; 95% CI, 41.0%–59.8%), and 65.0% (76 cases; 95% CI, 60.2%–78.5%), respectively.
The sensitivity of real-time PCR was much higher than C. difficile culture and the toxin immunoassay (P<0.005). Sigmoidoscopy was performed in only 16 cases and showed an appearance of antibiotic-associated colitis in 13 cases. Among these 16 cases, the diagnostic accuracy of sigmoidoscopy was 81.3% (95% CI, 67.8%–100%), which was lower than the diagnostic accuracy of real-time PCR (92.7%; 95% CI, 89.3%–96%).
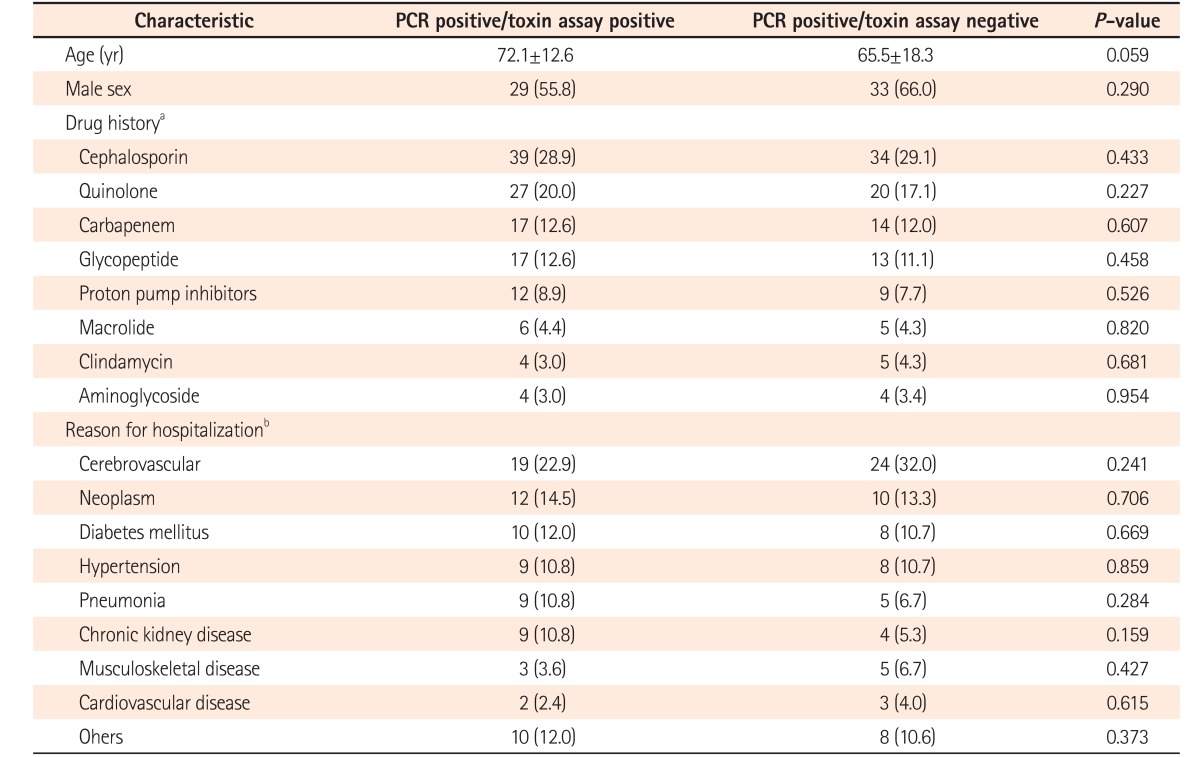
A total of 52 cases (44.4%) were diagnosed with CDI by both positive findings of real-time PCR and toxin A & B assay. Whereas 50 cases (42.7%) were diagnosed as CDI by real-time PCR positive, despite the toxin A & B assay being negative. In addition, 38 cases (32.5%) were diagnosed with CDI by real-time PCR positive although culture for C. difficile were negative. Notably, 34 cases (29.0%) were diagnosed with CDI by real-time PCR only (Fig. 1). When comparing the 2 groups regarding their clinical characteristics, there was no significant difference between 2 groups (Table 3).
In our study, 2 cases (1.7%) of hypervirulent CDI strain (027/NAP1/BI) were reported by real-time PCR method. One case had hypertension, diabetes mellitus, and end-stage renal disease as underlying diseases and developed CDI in the hospital after 49 days of stay. The patient was being treated for pneumonia and CDI in the intensive care unit. However, the patient expired due to septic shock with multi-organ failure. The other patient had gait disturbance and ventriculoperitoneal shunt owing to hydrocephalus. The patient was hospitalized for 38 days, before being ultimately discharged in a good condition. In both cases, oral vancomycin was administered to treat the CDI.
A recent report has suggested that the real-time PCR method is a more sensitive and accurate method for the diagnosis of CDI.9 Furthermore, a recent study observed a sensitivity of 99.1% for PCR, 83.3% for the toxin assay, and 51% for the culture method.10 In a European study, the sensitivity was reported to be 100% for real-time PCR and 58.8% for the toxin assay by ELISA.11 This study was performed in a tertiary teaching hospital and samples were collected prospectively from 150 hospitalized adult patients. The researchers suggested that real-time PCR method was a more sensitive and accurate method for the diagnosis of CDI and our study observed similar results. In the present study, different methods for detecting toxins were employed, which revealed that real-time PCR is the most sensitive method for the detection of toxin-producing C. difficile. In addition, we obtained meaningful results in which 34 cases (29.0%) were diagnosed with CDI by real-time PCR only. This finding suggested that the real-time PCR method could reduce the repeated exam of toxin assay or culture and make the rapid diagnosis of CDI, thereby reducing the duration of hospitalization stay and economic burden.
Regarding the time to obtain results, real-time PCR took 2.27 hours (136.62 minutes), toxin assay took 83.67 hours (5,020.66 minutes), and culture took 105.79 hours (6,347.68 minutes), respectively. This confirmed that real-time PCR results can be obtained more rapidly compared to the other methods tested, with a statistically significant difference (P<0.005). It has to be noted that this time is only measured from the beginning of the test to when the results are obtained. Therefore, the total time taken to complete the testing may differ from one test to the other. Although many studies have investigated the accuracy of CDI diagnostic methods, few studies have tested the time required to obtain results for CDI diagnosis. For example, a previous study reported that a cytotoxin assay needs 24 to 48 hours and stool culture requires 2 to 5 days.12 Furthermore, studies showed that the mean time until results could be reported to the wards was 1.53 hours for real-time PCR, 22.45 hours for positive culture cytotoxicity neutralization assay, 46.54 hours for negative culture cytotoxicity neutralization assay, and 4.47 hours for toxin enzyme immunoassay.13
14
We compared the characteristics of patients between the 2 groups of real-time PCR positive/toxin assay positive and real-time PCR positive/toxin assay negative. However, there was no significant difference between the 2 groups.
In our study, 2 cases of the hypervirulent CDI strain (027/NAP1/BI) were reported using the real-time PCR method. Ribotype 027 strain had toxin A, toxin B, and binary toxin with a deletion of the toxin regulator gene (tcdC).15
16 The first case of C. difficile ribotype 027 in Korea was reported in 2009.17 After that, 7 cases were reported at the tertiary-care hospital in Korea and there was a warning of a possible outbreak of CDI.18
19 In terms of detecting the hypervirulent CDI, the real-time PCR method possesses a benefits compared to other methods.
The economic effect of early diagnosis by real-time PCR has been debated. In a study by the University of Wisconsin, it has been reported that the rapid reporting of real-time PCR test results was associated with fewer diagnostic laboratory tests performed per patient and a shorter length of empiric antibiotic therapy.20 We also suggested that real-time PCR method is cost-effective in treating CDI patients. Real-time PCR is generally expensive relative to other methods; however, it can reduce the diagnostic delay of CDI. Therefore, real-time PCR can potentially lead to improved outcomes of CDI, which can result in significant hospital cost savings.21
Our study has several limitations. First, this study was conducted retrospectively with a relatively small sample size (n=117). Second, the time to obtain results was calculated from the beginning of the test to when the results were obtained, therefore this time may differ between hospitals depending on their reporting system.
In conclusion, we confirmed that the real-time PCR is superior to other diagnostic methods for CDI in terms of accuracy and rapidity. In contrast to other methods, it also helps to diagnose hypervirulent CDI, such as ribotype 027 infection.
References
1. O'Connor JR, Johnson S, Gerding DN. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology. 2009; 136:1913–1924. PMID: 19457419.
2. Pépin J, Valiquette L, Alary ME, et al. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ. 2004; 171:466–472. PMID: 15337727.

3. Ricciardi R, Rothenberger DA, Madoff RD, Baxter NN. Increasing prevalence and severity of Clostridium difficile colitis in hospitalized patients in the United States. Arch Surg. 2007; 142:624–631. PMID: 17638799.

4. Burke KE, Lamont JT. Clostridium difficile infection: a worldwide disease. Gut Liver. 2014; 8:1–6. PMID: 24516694.
5. Kim YS, Han DS, Kim YH, et al. Incidence and clinical features of Clostridium difficile infection in Korea: a nationwide study. Epidemiol Infect. 2013; 141:189–194. PMID: 22717061.

6. Doh YS, Kim YS, Jung HJ, et al. Long-term clinical outcome of Clostridium difficile infection in hospitalized patients: a single center study. Intest Res. 2014; 12:299–305. PMID: 25374496.

7. Yoon SY, Jung SA, Na SK, et al. What's the clinical features of colitis in elderly people in long-term care facilities? Intest Res. 2015; 13:128–134. PMID: 25931997.

8. Mueller S, Saunier K, Hanisch C, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006; 72:1027–1033. PMID: 16461645.

9. Barbut F, Monot M, Rousseau A, et al. Rapid diagnosis of Clostridium difficile infection by multiplex real-time PCR. Eur J Clin Microbiol Infect Dis. 2011; 30:1279–1285. PMID: 21487764.

10. Berry N, Sewell B, Jafri S, et al. Real-time polymerase chain reaction correlates well with clinical diagnosis of Clostridium difficile infection. J Hosp Infect. 2014; 87:109–114. PMID: 24795170.

11. de Jong E, de Jong AS, Bartels CJ, van der Rijt-van den Biggelaar C, Melchers WJ, Sturm PD. Clinical and laboratory evaluation of a real-time PCR for Clostridium difficile toxin A and B genes. Eur J Clin Microbiol Infect Dis. 2012; 31:2219–2225. PMID: 22327373.

12. Poutanen SM, Simor AE. Clostridium difficile-associated diarrhea in adults. CMAJ. 2006; 171:51–58.

13. Sewell B, Rees E, Thomas I, Ch’ng CL, Isaac M, Berry N. Cost and impact on patient length of stay of rapid molecular testing for Clostridium difficile. Infect Dis Ther. 2014; 3:281–293. PMID: 25183400.

14. Vaishnavi C. Clinical spectrum & pathogenesis of Clostridium difficile associated diseases. Indian J Med Res. 2010; 131:487–499. PMID: 20424299.
15. Goudarzi M, Seyedjavadi SS, Goudarzi H, Mehdizadeh Aghdam E, Nazeri S. Clostridium difficile infection: epidemiology, pathogenesis, risk factors, and therapeutic options. Scientifica (Cairo). 2014; 2014:916826. PMID: 24991448.
16. Warny M, Pepin J, Fang A, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005; 366:1079–1084. PMID: 16182895.

17. Tae CH, Jung SA, Song HJ, et al. The first case of antibiotic-associated colitis by Clostridium difficile PCR ribotype 027 in Korea. J Korean Med Sci. 2009; 24:520–524. PMID: 19543521.

18. Kim J, Kang JO, Kim H, et al. Epidemiology of Clostridium difficile infections in a tertiary-care hospital in Korea. Clin Microbiol Infect. 2013; 19:521–527. PMID: 22712697.

19. Kim H, Lee Y, Moon HW, Lim CS, Lee K, Chong Y. Emergence of Clostridium difficile ribotype 027 in Korea. Korean J Lab Med. 2011; 31:191–196. PMID: 21779194.
20. Peppard WJ, Ledeboer NA. Implementation of polymerase chain reaction to rule out Clostridium difficile infection is associated with reduced empiric antibiotic duration of therapy. Hosp Pharm. 2014; 49:639–643. PMID: 25477583.

21. Catanzaro M, Cirone J. Real-time polymerase chain reaction testing for Clostridium difficile reduces isolation time and improves patient management in a small community hospital. Am J Infect Control. 2012; 40:663–666. PMID: 22153847.

Fig. 1
Study process. A total of 207 cases from 116 patients were initially enrolled in this study and from these, 117 cases from 90 patients were diagnosed as Clostridium difficile infection (CDI).
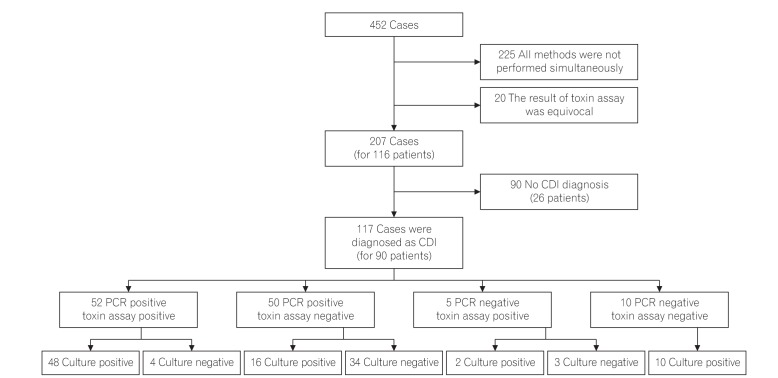
Table 3
Characteristics of Patients with Toxin Assay-Positive and -Negative Results with PCR Positive Results
Values are presented as median±SD or number (%).
aSome patients were prescribed more than 1 drug. Therefore, a total of 252 drugs was prescribed in the groups of real-time PCR positive/toxin assay positive and real-time PCR positive/toxin assay negative.
bA total of 158 underlying diseases were counted, of which some of the patients had more than 1 underlying disease in the real-time PCR positive/toxin assay positive and real-time PCR positive/toxin assay negative groups.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download