Abstract
Periodontitis is a common oral disease that is characterized by infection and inflammation of the tooth supporting tissues. While its incidence is highly associated with outgrowth of the pathogenic microbiome, some patients show signs of predisposition and quickly fall into recurrence after treatment. Recent research using genetic associations of candidates as well as genome-wide analysis highlights that variations in genes related to the inflammatory response are associated with an increased risk of periodontitis. Intriguingly, some of the genes are regulated by epigenetic modifications, supposedly established and reprogrammed in response to environmental stimuli. In addition, the treatment with epigenetic drugs improves treatment of periodontitis in a mouse model. In this review, we highlight some of the recent progress identifying genetic factors associated with periodontitis and point to promising approaches in epigenetic research that may contribute to the understanding of molecular mechanisms involving different responses in individuals and the early detection of predispositions that may guide in future oral treatment and disease prevention.
Periodontitis is one of the most common oral diseases in adult populations worldwide and is a major public health concern due to its substantial cost to the medical care system [1]. It is characterized by inflammation and destruction of tooth supporting tissues, in severe cases leading to tooth loss [2]. It is also highly associated with systemic inflammation, resulting in an increased risk for subsequent chronic diseases, such as cardiovascular diseases [3,4], diabetes [5], metabolic syndrome [6-8], pneumonia [9,10], and rheumatoid arthritis [11].
Periodontitis is a complex disease with an etiology involving multiple factors. It includes both extrinsic (modifiable) and intrinsic (nonmodifiable) factors. Although the definitive mechanisms remain unclear, inflammation and infection via outgrowth of multiple opportunistic microbes in the oral environment, including Porphyromonas gingivalis, Tannerella forsythia, and Actinobacillus actinomycetemcomitans, are a contributing factor. [12]. While the presence of microbiological pathogens is a factor leading to this condition, it is not solely sufficient to cause periodontitis. It was recently proposed that dysbiosis of the oral microbiota leads to periodontitis via interference of the host-microbial homeostasis, rather than simple outgrowths of a few pathogens [13,14]. Lifestyle-related factors, such as smoking and dietary patterns, as well as oral hygiene, have also been highly correlated with the prevalence of periodontitis [15,16]. While periodontal research so far has focused on studies of microbiological pathogenesis and oral environments, it is now widely accepted that susceptibility to inflammation is also determined by intrinsic factors such as genetics (Fig. 1A). This is emphasized by the fact that some people are more disease-susceptible or treatment resistant. In this review, we summarize the genetic factors that are of importance in the establishment and progression of periodontitis, and highlight what epigenetic alterations may be critical in the etiology of periodontitis, especially as key mediators between genetic and environmental factors. This information will provide future guidelines for development of novel biomarkers that will aid in the diagnosis, treatment, and cure of this common disease.
While genetic factors undoubtedly are very important in the development of periodontitis, genetic variations can only become a risk factor when challenged by extrinsic agents and physical insults. Genetic variations linked to complex diseases are not easily identified in multifactorial traits. Single nucleotide polymorphisms (SNP) of the DNA are often used as genetic markers when they can be linked to a distinct phenotype. Per definition, a SNP has to occur in at least 1% of a given population, while individual mutations, not fixed in the population, are referred to as single nucleotide variations (SNVs). Some SNPs alter gene expression levels that may influence host response levels to microbiological growth. For example, SNPs in receptors, antigen sensors in cell surfaces, and cytokines and chemokines have been shown to influence host immunity and inflammatory response [17,18]. Putative periodontitis-related SNPs have been investigated in the Fc-γ receptor (FCGR2A) [19-22], interleukin-1 (IL-1) [23,24], IL-4 [25-27], IL-6 [27,28], IL-10 [29,30], IL-18 [31,32], tumor necrosis factor alpha (TNFA) [23,33,34], vitamin D receptor [21,35-37], cluster of differentiation-14 [38-40], matrix metalloproteinase-1 [41,42], Toll-like receptor-2 (TLR-2) [43,44], TLR-4 [31,32,39,40,43,44], and cyclo-oxygenase-2 (COX-2) [45,46]. These studies suggest a connection between genetic variation and periodontitis at some loci, while their penetrance remains elusive. It is worth emphasizing that the connection between SNPs and periodontitis is not always strong because of variations within populations and subtypes of periodontitis.
Lately, multiple "omics" technologies have been utilized to increase the understanding of periodontitis disease progression by overcoming the limitations that candidate approaches have provided. The most prominent approaches are genomics, transcriptomics, proteomics, and metabolomics of the host as well as metagenomics of the oral microbiota [47] (Fig. 1B). From a large set of SNPs distributed over all chromosomes, genome-wide association studies (GWAS) have been applied comprehensively to identify genetic variations that are associated with periodontitis [48,49]. GWAS studies have identified novel genes for susceptibility, including GLT6D1 in aggressive periodontitis [48], NIN, NPY, and WNT5A in severe chronic periodontitis, and NCR2 and EMR1 in moderate chronic periodontitis [49]. While it has not been fully determined how the identified susceptibility genes affect pathogenesis, GWAS has provided novel insights into the etiology of periodontitis genome-wide, compared to previously taken candidate approaches.
Furthermore, apart from genome-wide genetic variation analysis, total gene transcript analysis, generally referred to as transcriptomics, has been conducted on periodontal tissues as well as peripheral blood cells [50-52]. Proteomics and metabolomics have also been applied to saliva or gingival crevicular fluid to identify proteins and metabolites with a negative or positive influence on host defense mechanisms [53-55]. It is noteworthy that the levels of transcripts, proteins and metabolites may reflect not only the genetic programming, but also the consequences of response to environmental factors and disease progression. There are layers of chemical modifications on the DNA and its associated proteins that regulate gene expression, commonly referred to as epigenetic effects. These become established or erased based on an environmental response, subsequently leading to intracellular signaling.
The genetic material, thought of as a database of cellular information, is not only charged by its coding capacity. The last three decades of genetic research has uncovered key factors that reproducibly bind the DNA and organize it into functional units [56]. The DNA is wrapped around histone proteins, the nucleosomes, which serve the purpose of condensing and decondensing the DNA depending on gene activity, and in maintaining chromosome integrity at times of cessation and cell division. The nucleosomes are fairly evenly distributed over the chromosomes, and there are recently developed techniques that allow us to accurately predict where those are positioned [57]. Importantly, each protein in the nucleosome is subject to post-translational modifications, appearing at defined positions of the genome, and is strongly correlated with the activities observed at the DNA (e.g., transcriptional activity and elongation). For instance, trimethylation at histone H3 lysine 4 (H3K4me3) is associated with transcriptional activation, while H3K9me3 and H3K27me3 are associated with transcriptional repression [58] (Fig. 2). The most well characterized epigenetic modification is DNA methylation, which occurs at the 5th carbon of cytosines in mammals (5mC), most commonly next to a guanosine (CpG). Methyl-groups are placed by DNA methyltransferases (DNMTs) that catalyze the transfer from the methyl donor S-adenosyl methionine to the cytosines. Methylation at previously unmethylated sites is placed by the DNMT3a/b, while it is commonly maintained by the DNMT1, referred to as the de novo and maintenance methyltransferases, respectively. Together they assure that the vast majority of the genome is methylated at all times, leaving only regulatory elements like promoters and enhancers, and CpG-rich islands unmethylated [59]. The acquisition of DNA methylation at the promoter is predominantly associated with gene silencing (Fig. 2). DNA methylation can be further modified via oxidation by the TET proteins, implicated in DNA demethylation processes via the base excision repair machinery [60]. On many occasions, multiple epigenetic modifications, including DNA methylation and various histone modifications, work coordinately or antagonistically [59,61,62].
It is easy to conceive that misplaced modifications and the reduced dynamics of their distribution lead to obstructed gene activity and disease (Fig. 3). This generally occurs as a result of mutations of factors that directly make or influence epigenetic modifications [63]. Since each factor has an influence on a genome-wide level, the effect can be dramatic.
There is an ever increasing amount of evidence showing that understanding the epigenetic pattern in disease progression will provide invaluable information in the diagnosis and treatment of human disease [64]. Genome-wide analysis of epigenetic patterns in tissues that undergo defined changes as a result of external stress have provided insight into how cells respond to external factors [65]. Furthermore, such analysis offers detailed insight into how the cells try to cope with the changes and what may be the outstanding factors that determine whether the changes can be dealt with or result in skewed differentiation or cell death.
The response is hidden in the genetic background, since the results of GWAS have identified correlations with disease susceptibility and progression [66]. This was investigated by screening for quantitative trait loci that are in linkage disequilibrium with genes in close proximity and inferred to be causative in disease predisposition. Extensive studies have been conducted to identify gene expression patterns and/or epigenetically modified loci to determine which ones are correlated with a particular disease [67,68].
An important part of identifying and characterizing the epigenetic pattern in development and disease is to use the information to predict the treatment and cure of diseases based on a suggested genetic response [69]. This information has led to the identification of biomarkers that directly correlate with a defined condition [70]. Similarly, epigenetic patterns that suggest a predisposition for a particular disease have been identified and should be possible to use as the basis for developing personalized and preventive treatment regimes to prevent future problems. However, this is more a vision than a reality in today's medicine.
The placement of epigenetic modifications is tightly controlled both spatially and temporally. Each tissue has a unique epigenetic profile, and changes do occur as a result of developmental and regenerative processes. There is clear evidence that embryonic stem cells have a unique epigenetic pattern that changes upon differentiational cues [71]. Extrinsic factors, such as hormones, regulate differentiation, and in effect influence epigenetic modifications [63]. The epigenetic pattern that we observe in any particular tissue at any particular point in time is a reflection of its activity [72]. Most of the information is then further reflected by its gene expression pattern. Hence, the observed epigenetic pattern can be used to infer the transcriptional condition of the cell or tissue.
Treatment of cells in vitro generates a defined epigenetic pattern, as evidenced by studies of induced pluripotent stem cells [73] and epithelial-to-mesenchymal transition [74]. The gut microbiome can alter the epigenetic pattern of the gut endothelial cells [75,76] by excreting signals that trigger a response. Likewise, a similar effect can be achieved in the oral cavity, which is under the constant influence of extrinsic factors and foreign agents from food intake. Oral hygiene is naturally a contributing factor to oral health. Mounting evidence suggest that a lifestyle of smoking, food intake, lack of exercise, and use of drugs strongly influences the epigenetic pattern and predisposition to most conditions that lead to human disease [77].
A typical inflammatory response results in the upregulation of genes associated with the production of lectins that then coat epithelial cell surfaces, with the function of recruiting neutrophiles to the site of infection. This initiates an immune response that involves both innate and adaptive-related processes. It is at this stage that epigenetic regulation of gene expression patterns seems to play the most important role [78] and is key in the upregulation of proinflammatory cytokines and other signaling molecules to activate a full response from immune cells, while simultaneously downregulating anti-inflammatory cytokines. The cytokine genes have been suggested as targets of multiple epigenetic events including transcriptional activation via loss of DNA methylation and active histone modifications at regulatory elements [79-81].
The IL-1, IL-2, IL-6, IL-8, IL-10, and IL-12 genes may be regulated by epigenetic mechanisms [79,82]. In chronic obstructive pulmonary disease, proinflammatory cytokines (IL-1, IL-2, IL-8, and IL-12) are highly expressed via increased H3K9 acetylation at the promoters of CBP/p300 and decreased histone deacetylase activity, following the recruitment of NF-kB to gene promoters [83]. TNFA, encoding TNF-α, is also regulated by epigenetic modifications both constitutively and in response to acute stimulation in myeloid cells [84]. DNA methylation also involves cytokine expression such as interferon gamma (IFN)-γ and IL-10 by transcriptional inactivation and skewed differentiation toward IL-10-expressing regulatory T cells, respectively [85,86]. In addition to cytokines, TLR-2 and TLR-4, associated with an increased proinflammatory response, are regulated by DNA methylation in bronchial and intestinal epithelial cells [87,88]. The TLRs, expressed on the cell surface, are involved in the recognition of bacterial components such as lipoproteins, lipo-polysaccharide, flagellin, and DNA [89], so that DNA methylation-mediated regulation of their expression is crucial to determining the magnitude of the bacteria-induced response.
Intriguingly, inflammation signaling itself influences epigenetic changes in cells. IL-6 and IL-1b promote transcription or protein activity of DNMTs, respectively [90,91]. Cytokine-induced methylation changes lead to transcriptional repression of multiple target genes. Taken together, epigenetic mechanisms play a key role in the initiation and progression of inflammation by determination of cytokine profiles in response to environmental stimuli, but also by regulating downstream target genes in response to cytokines.
Epigenetic studies on the epithelial lining of the oral cavity are in their infancy, but several studies suggest that these cells have a unique capacity to respond to environmental factors. In the periodontal cavity, the inflammatory response involves upregulation of transcription factors (e.g., NF-κB and STAT) and epigenetic chromatin changes similar to other inflammatory diseases [92] (Table 1). Chronic periodontitis patients showed overexpression of cytokines such as IL-6 and IFN-γ in their inflamed tissues [28,93]. The associations between IL-6 and periodontitis are also supported by genetic evidence [27,28]. The expression changes of some loci (e.g., IFNG), occur as a result of the loss of methylation at their promoters [93]. On the other hand, the overexpression of IL-6 is not associated with DNA methylation at its promoter. IL-6 upregulation may rather activate the DNMTs [90], leading to methylation changes at the IL-6-induced target genes and development of a chronic inflammatory condition.
Recently, Zhang et al. [94] showed that the TNFA promoter was hypermethylated at two CpG sites, resulting in decreased expression. By reversing the methylation by treatment with a demethylating agent in vitro, it caused increased expression of TNFA, indicating that the methylation indeed regulated the expression. Lower expression in patients compared to healthy controls was, however, in conflict with a previous report [95]. The authors speculated that the discrepancy might be due to the difference in the state of inflammation of the patients, considering the fact that only severely afflicted patients showed elevated TNF-α [96]. It could also be due to the not-always-direct relationship between the mRNA level and protein level. Either way, further investigations are required to determine the role of TNF-α in periodontitis.
Further evidence of epigenetic changes associated with periodontitis comes from data on COX-2, an enzyme governing the production of prostaglandins that promote inflammation and pain. It has been reported that COX-2 inhibitors were able to reduce the symptoms of periodontitis patients [97]. Nevertheless, COX-2 expression in inflamed gingival tissues from chronic periodontitis patients was lower and its promoter was hypermethylated [98], which was confirmed by an independent study [99]. Similar to TNF-α, methylation changes occur more frequently in periodontitis than in healthy individuals, but it remains unclear whether it is linked to periodontitis etiology or rather indicates the consequence of DNMT activation by persistent chronic inflammation.
In addition to DNA methylation, other epigenetic changes such as histone modifications are involved in periodontitis. Treatment by HDAC inhibitors efficiently suppressed periodontal bone loss in a mouse model of periodontitis [100]. Treatment with novel HDAC inhibitors, such as 1179.4b and MS-275, on P. gingivalis-inoculated mice resulted in significantly reduced bone loss, indicating that maintenance of acetylation is crucial to preventing bone loss.
Collectively, gingival tissues from periodontitis patients seem to have altered epigenetic patterns, particularly at inflammation-related genes. However, it needs to be determined whether 1) the alterations account for the susceptibility like genetic variations in those loci; 2) they are directly related to a mechanism driving the pathogenesis by transcriptional changes in critical target genes; or 3) they are just consequences of chronic inflammatory events. Future genome-wide studies on epigenetic factors promise to provide insightful answers to these questions.
The understanding of periodontitis has substantially benefited from the recent identification of genetic factors (in particular SNPs and SNVs) and epigenetic regulatory mechanisms (i.e., aberrant epigenetic patterns). These findings provide novel insight into the etiology of periodontitis, especially regarding the tissue response to infection, as well as highlighting putative mechanisms by which genetic and environmental factors influence each other. While analysis of candidate inflammation-related genetic factors have been common so far, current ongoing genome-wide analysis of genetic variation and epigenetic alterations in periodontitis will likely expand our understanding of the pathogenesis of periodontitis in an unbiased way. The newly gathered information will be used in developing novel therapeutic interventions, potentially involving epigenetic modifiers, leading the way to personalized medicinal treatment and preventional regimes.
Figures and Tables
Figure 1
Intrinsic and extrinsic risk factors for periodontitis. (A) Factors influencing the pathogenesis of periodontitis in the oral cavity. (B) Methods of large-scale analysis to identify genetic factors, epigenetic patterns, comprehensive transcriptomics, proteomics, metabolomics, and microbiomics in close connection to environmental factors.
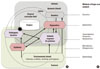
Figure 2
Chromatin changes by epigenetic modifications and transcriptional status. Schematic overview of chromatin changes by typical epigenetic modifications. Combinations of epigenetic modifications contribute to determining chromatin structure, leading to an open or closed chromatin configuration and transcriptional state. For example, reduced DNA methylation at the promoter of the interferon gamma (IFNG) gene is associated with increased expression of IFNG in the inflamed tissues from periodontitis patients, compared to healthy periodontal tissues.
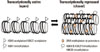
Figure 3
Genetic and epigenetic alterations in disease progression. Genetic and epigenetic alterations contribute to gene expression either with or without changes in DNA sequences, respectively. Normal expression can be interrupted via genetic alteration by production of abnormal protein or altered efficiency of gene transcription. Likewise, interruption can be accomplished by epigenetic alterations at transcriptionally regulatory regions. The 'black box' represents exons while the 'grey box' represents introns or regulatory regions. The 'highlighted G' represents a nucleotide that has replaced a dominant or a normal nucleotide as genetic variation or mutation, respectively. White and black circles indicate the different statuses of epigenetic modifications at the regulatory elements of a given gene. Specifically, white circles indicate unmethylated cytosines at the promoter that usually allow active transcription, while 'black circles' indicate methylated cytosines at the promoter that usually suppress transcription.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. 2012018819).
References
1. Petersen PE, Ogawa H. The global burden of periodontal disease: towards integration with chronic disease prevention and control. Periodontol 2000. 2012; 60:15–39.

2. Petersen PE, Ogawa H. Strengthening the prevention of periodontal disease: the WHO approach. J Periodontol. 2005; 76:2187–2193.

3. Genco RJ, Van Dyke TE. Prevention: reducing the risk of CVD in patients with periodontitis. Nat Rev Cardiol. 2010; 7:479–480.

4. Tonetti MS. Periodontitis and risk for atherosclerosis: an update on intervention trials. J Clin Periodontol. 2009; 36:Suppl 10. 15–19.

5. Lalla E, Papapanou PN. Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat Rev Endocrinol. 2011; 7:738–748.

6. Nibali L, Tatarakis N, Needleman I, Tu YK, D'Aiuto F, Rizzo M, et al. Clinical review: Association between metabolic syndrome and periodontitis: a systematic review and meta-analysis. J Clin Endocrinol Metab. 2013; 98:913–920.

7. Han DH, Lim SY, Sun BC, Paek D, Kim HD. The association of metabolic syndrome with periodontal disease is confounded by age and smoking in a Korean population: the Shiwha-Banwol Environmental Health Study. J Clin Periodontol. 2010; 37:609–616.

8. Kwon YE, Ha JE, Paik DI, Jin BH, Bae KH. The relationship between periodontitis and metabolic syndrome among a Korean nationally representative sample of adults. J Clin Periodontol. 2011; 38:781–786.

9. Awano S, Ansai T, Takata Y, Soh I, Akifusa S, Hamasaki T, et al. Oral health and mortality risk from pneumonia in the elderly. J Dent Res. 2008; 87:334–339.

10. Paju S, Scannapieco FA. Oral biofilms, periodontitis, and pulmonary infections. Oral Dis. 2007; 13:508–512.

11. Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA-the citrullinated enolase connection. Nat Rev Rheumatol. 2010; 6:727–730.

13. Hajishengallis G, Lambris JD. Complement and dysbiosis in periodontal disease. Immunobiology. 2012; 217:1111–1116.

14. Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012; 91:816–820.

15. Van Dyke TE, Sheilesh D. Risk factors for periodontitis. J Int Acad Periodontol. 2005; 7:3–7.
16. Van der Velden U, Abbas F, Armand S, Loos BG, Timmerman MF, Van der Weijden GA, et al. Java project on periodontal diseases. The natural development of periodontitis: risk factors, risk predictors and risk determinants. J Clin Periodontol. 2006; 33:540–548.

17. Yamamoto K, Kobayashi T, Grossi S, Ho AW, Genco RJ, Yoshie H, et al. Association of Fcgamma receptor IIa genotype with chronic periodontitis in Caucasians. J Periodontol. 2004; 75:517–522.

18. Fu Y, Korostoff JM, Fine DH, Wilson ME. Fc gamma receptor genes as risk markers for localized aggressive periodontitis in African-Americans. J Periodontol. 2002; 73:517–523.

19. Yoshihara A, Sugita N, Yamamoto K, Kobayashi T, Miyazaki H, Yoshi H. Analysis of vitamin D and Fcgamma receptor polymorphisms in Japanese patients with generalized early-onset periodontitis. J Dent Res. 2001; 80:2051–2054.

20. Chai L, Song YQ, Zee KY, Leung WK. SNPs of Fc-gamma receptor genes and chronic periodontitis. J Dent Res. 2010; 89:705–710.

21. Maria de Freitas N, Imbronito AV, Neves AC, Nunes FD, Pustiglioni FE, Lotufo RF. Analysis of IL-1A(-889) and TNFA(-308) gene polymorphism in Brazilian patients with generalized aggressive periodontitis. Eur Cytokine Netw. 2007; 18:142–147.
22. Komatsu Y, Galicia JC, Kobayashi T, Yamazaki K, Yoshie H. Association of interleukin-1 receptor antagonist +2018 gene polymorphism with Japanese chronic periodontitis patients using a novel genotyping method. Int J Immunogenet. 2008; 35:165–170.

23. Kang BY, Choi YK, Choi WH, Kim KT, Choi SS, Kim K, et al. Two polymorphisms of interleukin-4 gene in Korean adult periodontitis. Arch Pharm Res. 2003; 26:482–486.

24. Michel J, Gonzales JR, Wunderlich D, Diete A, Herrmann JM, Meyle J. Interleukin-4 polymorphisms in early onset periodontitis. J Clin Periodontol. 2001; 28:483–488.

25. Aoyagi T, Sugawara-Aoyagi M, Yamazaki K, Hara K. Interleukin 4 (IL-4) and IL-6-producing memory T-cells in peripheral blood and gingival tissue in periodontitis patients with high serum antibody titers to Porphyromonas gingivalis. Oral Microbiol Immunol. 1995; 10:304–310.

26. Stefani FA, Viana MB, Dupim AC, Brito JA, Gomez RS, da Costa JE, et al. Expression, polymorphism and methylation pattern of interleukin-6 in periodontal tissues. Immunobiology. 2013; 218:1012–1017.

27. Mellati E, Arab HR, Tavakkol-Afshari J, Ebadian AR, Radvar M. Analysis of -1082 IL-10 gene polymorphism in Iranian patients with generalized aggressive periodontitis. Med Sci Monit. 2007; 13:CR510–CR514.
28. Babel N, Cherepnev G, Babel D, Tropmann A, Hammer M, Volk HD, et al. Analysis of tumor necrosis factor-alpha, transforming growth factor-beta, interleukin-10, IL-6, and interferon-gamma gene polymorphisms in patients with chronic periodontitis. J Periodontol. 2006; 77:1978–1983.

29. Noack B, Gorgens H, Lorenz K, Ziegler A, Hoffmann T, Schackert HK. TLR4 and IL-18 gene variants in aggressive periodontitis. J Clin Periodontol. 2008; 35:1020–1026.
30. Noack B, Gorgens H, Lorenz K, Schackert HK, Hoffmann T. TLR4 and IL-18 gene variants in chronic periodontitis: impact on disease susceptibility and severity. Immunol Invest. 2009; 38:297–310.

31. Fassmann A, Holla LI, Buckova D, Vasku A, Znojil V, Vanek J. Polymorphisms in the +252(A/G) lymphotoxin-alpha and the -308(A/G) tumor necrosis factor-alpha genes and susceptibility to chronic periodontitis in a Czech population. J Periodontal Res. 2003; 38:394–399.

32. Garlet GP, Trombone AP, Menezes R, Letra A, Repeke CE, Vieira AE, et al. The use of chronic gingivitis as reference status increases the power and odds of periodontitis genetic studies: a proposal based in the exposure concept and clearer resistance and susceptibility phenotypes definition. J Clin Periodontol. 2012; 39:323–332.

33. Deng H, Liu F, Pan Y, Jin X, Wang H, Cao J. BsmI, TaqI, ApaI, and FokI polymorphisms in the vitamin D receptor gene and periodontitis: a meta-analysis of 15 studies including 1338 cases and 1302 controls. J Clin Periodontol. 2011; 38:199–207.

34. Park KS, Nam JH, Choi J. The short vitamin D receptor is associated with increased risk for generalized aggressive periodontitis. J Clin Periodontol. 2006; 33:524–528.

35. Sun JL, Meng HX, Cao CF, Tachi Y, Shinohara M, Ueda M, et al. Relationship between vitamin D receptor gene polymorphism and periodontitis. J Periodontal Res. 2002; 37:263–267.

36. Nicu EA, Laine ML, Morre SA, Van der Velden U, Loos BG. Soluble CD14 in periodontitis. Innate Immun. 2009; 15:121–128.

37. Laine ML, Morre SA, Murillo LS, van Winkelhoff AJ, Peña AS. CD14 and TLR4 gene polymorphisms in adult periodontitis. J Dent Res. 2005; 84:1042–1046.

38. James JA, Poulton KV, Haworth SE, Payne D, McKay IJ, Clarke FM, et al. Polymorphisms of TLR4 but not CD14 are associated with a decreased risk of aggressive periodontitis. J Clin Periodontol. 2007; 34:111–117.

39. Holla LI, Jurajda M, Fassmann A, Dvorakova N, Znojil V, Vacha J. Genetic variations in the matrix metalloproteinase-1 promoter and risk of susceptibility and/or severity of chronic periodontitis in the Czech population. J Clin Periodontol. 2004; 31:685–690.

40. Li D, Cai Q, Ma L, Wang M, Ma J, Zhang W, et al. Association between MMP-1 g.-1607dupG polymorphism and periodontitis susceptibility: a meta-analysis. PLoS One. 2013; 8:e59513.

41. Emingil G, Berdeli A, Baylas H, Saygan BH, Gürkan A, Köse T, et al. Toll-like receptor 2 and 4 gene polymorphisms in generalized aggressive periodontitis. J Periodontol. 2007; 78:1968–1977.

42. Berdeli A, Emingil G, Han Saygan B, Gurkan A, Atilla G, Köse T, et al. TLR2 Arg753Gly, TLR4 Asp299Gly and Thr399Ile gene polymorphisms are not associated with chronic periodontitis in a Turkish population. J Clin Periodontol. 2007; 34:551–557.

43. Daing A, Singh SV, Saimbi CS, Khan MA, Rath SK. Cyclooxygenase 2 gene polymorphisms and chronic periodontitis in a North Indian population: a pilot study. J Periodontal Implant Sci. 2012; 42:151–157.

44. Li Y, Xu L, Hasturk H, Kantarci A, DePalma SR, Van Dyke TE. Localized aggressive periodontitis is linked to human chromosome 1q25. Hum Genet. 2004; 114:291–297.

45. Chai L, Song YQ, Leung WK. Genetic polymorphism studies in periodontitis and Fcγ receptors. J Periodontal Res. 2012; 47:273–285.

46. Laine ML, Crielaard W, Loos BG. Genetic susceptibility to periodontitis. Periodontol 2000. 2012; 58:37–68.

47. Grant MM. What do 'omic technologies have to offer periodontal clinical practice in the future? J Periodontal Res. 2012; 47:2–14.

48. Schaefer AS, Richter GM, Nothnagel M, Manke T, Dommisch H, Jacobs G, et al. A genome-wide association study identifies GLT6D1 as a susceptibility locus for periodontitis. Hum Mol Genet. 2010; 19:553–562.

49. Divaris K, Monda KL, North KE, Olshan AF, Reynolds LM, Hsueh WC, et al. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Hum Mol Genet. 2013; 22:2312–2324.

50. Demmer RT, Behle JH, Wolf DL, Handfield M, Kebschull M, Celenti R, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008; 79:2112–2124.

51. Papapanou PN, Behle JH, Kebschull M, Celenti R, Wolf DL, Handfield M, et al. Subgingival bacterial colonization profiles correlate with gingival tissue gene expression. BMC Microbiol. 2009; 9:221.

52. Papapanou PN, Sedaghatfar MH, Demmer RT, Wolf DL, Yang J, Roth GA, et al. Periodontal therapy alters gene expression of peripheral blood monocytes. J Clin Periodontol. 2007; 34:736–747.

53. Kojima T, Andersen E, Sanchez JC, Wilkins MR, Hochstrasser DF, Pralong WF, et al. Human gingival crevicular fluid contains MRP8 (S100A8) and MRP14 (S100A9), two calcium-binding proteins of the S100 family. J Dent Res. 2000; 79:740–747.

54. Wu Y, Shu R, Luo LJ, Ge LH, Xie YF. Initial comparison of proteomic profiles of whole unstimulated saliva obtained from generalized aggressive periodontitis patients and healthy control subjects. J Periodontal Res. 2009; 44:636–644.

55. Barnes VM, Teles R, Trivedi HM, Devizio W, Xu T, Mitchell MW, et al. Acceleration of purine degradation by periodontal diseases. J Dent Res. 2009; 88:851–855.

56. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395.

57. Wei G, Hu G, Cui K, Zhao K. Genome-wide mapping of nucleosome occupancy, histone modifications, and gene expression using next-generation sequencing technology. Methods Enzymol. 2012; 513:297–313.

58. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837.

59. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012; 13:484–492.

60. Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012; 139:1895–1902.

61. Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008; 105:12979–12984.

62. Lindroth AM, Park YJ, McLean CM, Dokshin GA, Persson JM, Herman H, et al. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet. 2008; 4:e1000145.

63. Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012; 131:1565–1589.

64. Kilpinen H, Dermitzakis ET. Genetic and epigenetic contribution to complex traits. Hum Mol Genet. 2012; 21:R24–R28.

65. Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010; 330:612–616.

66. Tsai PC, Spector TD, Bell JT. Using epigenome-wide association scans of DNA methylation in age-related complex human traits. Epigenomics. 2012; 4:511–526.

68. Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010; 464:1351–1356.

70. How Kit A, Nielsen HM, Tost J. DNA methylation based biomarkers: practical considerations and applications. Biochimie. 2012; 94:2314–2337.

71. Mendenhall EM, Bernstein BE. Chromatin state maps: new technologies, new insights. Curr Opin Genet Dev. 2008; 18:109–115.

72. Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013; 14:204–220.

73. Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010; 28:1079–1088.

74. McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011; 18:867–874.

75. Kellermayer R. Epigenetics and the developmental origins of inflammatory bowel diseases. Can J Gastroenterol. 2012; 26:909–915.

76. Barros SP, Offenbacher S. Epigenetics: connecting environment and genotype to phenotype and disease. J Dent Res. 2009; 88:400–408.

77. Alegria-Torres JA, Baccarelli A, Bollati V. Epigenetics and lifestyle. Epigenomics. 2011; 3:267–277.

78. Wen H, Schaller MA, Dou Y, Hogaboam CM, Kunkel SL. Dendritic cells at the interface of innate and acquired immunity: the role for epigenetic changes. J Leukoc Biol. 2008; 83:439–446.

79. Fitzpatrick DR, Wilson CB. Methylation and demethylation in the regulation of genes, cells, and responses in the immune system. Clin Immunol. 2003; 109:37–45.

80. O'Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011; 11:239–250.
81. Nielsen HM, Tost J. Epigenetic changes in inflammatory and autoimmune diseases. Subcell Biochem. 2012; 61:455–478.

82. Villagra A, Sotomayor EM, Seto E. Histone deacetylases and the immunological network: implications in cancer and inflammation. Oncogene. 2010; 29:157–173.

83. Barnes PJ. Targeting the epigenome in the treatment of asthma and chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2009; 6:693–696.

84. Sullivan KE, Reddy AB, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, et al. Epigenetic regulation of tumor necrosis factor alpha. Mol Cell Biol. 2007; 27:5147–5160.

85. White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO- T cells. J Immunol. 2002; 168:2820–2827.

86. Adcock IM, Tsaprouni L, Bhavsar P, Ito K. Epigenetic regulation of airway inflammation. Curr Opin Immunol. 2007; 19:694–700.

87. Shuto T, Furuta T, Oba M, Xu H, Li JD, Cheung J, et al. Promoter hypomethylation of Toll-like receptor-2 gene is associated with increased proinflammatory response toward bacterial peptidoglycan in cystic fibrosis bronchial epithelial cells. FASEB J. 2006; 20:782–784.

88. Takahashi K, Sugi Y, Hosono A, Kaminogawa S. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J Immunol. 2009; 183:6522–6529.

89. Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. 2007; 13:552–559.

90. Hodge DR, Xiao W, Clausen PA, Heidecker G, Szyf M, Farrar WL. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem. 2001; 276:39508–39511.

91. Hmadcha A, Bedoya FJ, Sobrino F, Pintado E. Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production. J Exp Med. 1999; 190:1595–1604.

93. Zhang S, Crivello A, Offenbacher S, Moretti A, Paquette DW, Barros SP. Interferon-gamma promoter hypomethylation and increased expression in chronic periodontitis. J Clin Periodontol. 2010; 37:953–961.

94. Zhang S, Barros SP, Moretti AJ, Yu N, Zhou J, Preisser JS, et al. Epigenetic regulation of TNFA expression in periodontal disease. J Periodontol. 2013; 01. 31. [Epub]. http://dx.doi.org/10.1902/jop.2013.120294.
95. Gorska R, Gregorek H, Kowalski J, Laskus-Perendyk A, Syczewska M, Madalinski K. Relationship between clinical parameters and cytokine profiles in inflamed gingival tissue and serum samples from patients with chronic periodontitis. J Clin Periodontol. 2003; 30:1046–1052.

96. Salvi GE, Brown CE, Fujihashi K, Kiyono H, Smith FW, Beck JD, et al. Inflammatory mediators of the terminal dentition in adult and early onset periodontitis. J Periodontal Res. 1998; 33:212–225.

97. Pinho Mde N, Pereira LB, de Souza SL, Palioto DB, Grisi MF, Novaes AB Jr, et al. Short-term effect of COX-2 selective inhibitor as an adjunct for the treatment of periodontal disease: a clinical double-blind study in humans. Braz Dent J. 2008; 19:323–328.

98. Zhang S, Barros SP, Niculescu MD, Moretti AJ, Preisser JS, Offenbacher S. Alteration of PTGS2 promoter methylation in chronic periodontitis. J Dent Res. 2010; 89:133–137.

99. Loo WT, Jin L, Cheung MN, Wang M, Chow LW. Epigenetic change in E-cadherin and COX-2 to predict chronic periodontitis. J Transl Med. 2010; 8:110.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download