Abstract
Objectives
From the point of view of clinical data representation, this study attempted to identify obstacles in translating clinical narrative guidelines into computer interpretable format and integrating the guidelines with data in Electronic Health Records in China.
Methods
Based on SAGE and K4CARE formulism, a Chinese clinical practice guideline for hypertension was modeled in Protégé by building an ontology that had three components: flowchart, node, and vMR. Meanwhile, data items imperative in Electronic Health Records for patients with hypertension were reviewed and compared with those from the ontology so as to identify conflicts and gaps between.
Results
A set of flowcharts was built. A flowchart comprises three kinds of node: State, Decision, and Act, each has a set of attributes, including data input/output that exports data items, which then were specified following ClinicalStatement of HL7 vMR. A total of 140 data items were extracted from the ontology. In modeling the guideline, some narratives were found too inexplicit to formulate, and encoding data was quite difficult. Additionally, it was found in the healthcare records that there were 8 data items left out, and 10 data items defined differently compared to the extracted data items.
Conclusions
The obstacles in modeling a clinical guideline and integrating with data in Electronic Health Records include narrative ambiguity of the guideline, gaps and inconsistencies in representing some data items between the guideline and the patient' records, and unavailability of a unified medical coding system. Therefore, collaborations among various participants in developing guidelines and Electronic Health Record specifications is needed in China.
To encourage evidence-based practice, clinical decision support systems (CDSSs) that provide digitized clinical practice guidelines (CPGs) and critical pathways are being actively introduced into the medical field for the health professional's use in clinical settings [1,2]. CDSSs can also help people with chronic diseases manage their own health by providing instant knowledge and recommendations [3,4]. One of the key steps in developing a CPG-based CDSS is building computer interpretable guidelines (CIGs), also called guideline ontology through guideline representation languages, which define the declarative knowledge of complex medical pathways. Internationally, there are several languages and models to represent guidelines, such as GLIF3, SAGE, PROforma, etc. [5,6]. No matter what kind of techniques a CDSS uses, it delivers alerts, reminders, and suggestions based on not only decision rules specified in CPGs but also relevant patient data describing patient's health status collected by Electronic Health Record (EHR) systems [7]. Therefore, a large amount of routine clinical data needed by a CDSS has to be represented in an interoperable way with unambiguous meanings, and it must be consistent with those in CIGs so as to be retrieved and used by CDSS. To represent clinical data in a standard way for CDSSs, HL7 developed a data model called virtual medical record (vMR) based on the HL7 Reference Information Model (RIM) [8]. vMR represents clinical information inputs and outputs that can be exchanged between CDS engines and clinical information systems through mechanisms such as CDS services. The Domain Analysis Model (DAM) of vMR includes structural specifications for inputs and outputs of CDSS engines, which consists of several information classes, such as observation, encounter, problem, adverse reaction, goal, procedure, medication order, etc., each representing a specialization of the RIM Act class. vMR is employed by SAGE to support information communication between CDSSs and local clinical information systems [9].
Hypertension is a significant risk factor for people's health. It is estimated that there are currently 200 million hypertension patients in China [10,11]. To cope with the prevalence and the characteristic dangers of hypertension, the Chinese government has established a strategy of "focusing on prevention and transferring the healthcare downwards to primary health sectors." Consequently, hypertension has been one of major chronic diseases managed by primary healthcare organizations at the community level, and a clinical practice guideline specifically for primary healthcare of hypertension was developed in 2009, in which general knowledge about the disease was documented comprehensively to guide general practitioners to monitor, evaluate, medicate, and instruct hypertension patients [10]. Meanwhile, the national government imposed a set of requirements for the delivery of public health services and required that the data needed for chronic disease management, including hypertension, must be recorded by EHR systems in primary healthcare organizations for the purposes of evidence-based, continuing patient healthcare and public health policy making [12]. The data items and their formats in EHR systems that already exist or are implemented in primary healthcare facilities are also stated in the national specification.
Like many other clinical guidelines, the hypertension guideline has not been fully followed in daily healthcare practice. Several factors restricting physicians' adherence to clinical guidelines have been identified [13]. Investigations in other countries have revealed that data standardization is a critical factor for CDSS development [14,15,16,17]. In China, there are very few computerized guidelines to date in routine clinical use. Furthermore, there have not been investigations yet on whether the existing narrative guidelines in Chinese are able to be computerized or whether the data that information systems collected following the announcement of national specifications can meet the needs of CDSS implementation.
Therefore, this research tried to identify underlying obstacles in implementing clinical guidelines from the aspect of clinical data representation in the context of hypertension by investigating the medical statements in both the guideline and electronic patient record. Finally, we suggest some considerations that need to be taken into account in the development of clinical guidelines and EHR systems in China.
We modeled the hypertension guideline based on SAGE and K4CARE formalism [18], and took Protégé 3.4.3 as the modeling tool [19,20]. In representing related clinical data, we followed HL7 vMR DAM. The process is summarized as follows.
First, we extracted and formalized the narrative guideline as an ontology. The ontology has three successive components: flowchart, node, and vMR. There are three kinds of nodes in flowchart: State, Decision, and Act, which are related to and instantiated in node. The attributes of a node were set as label, description, data input/output, and vMR class. Data items were abstracted from the attributes of data input/output and organized into relative classes of vMR. Because data items are instances of the classes they belong to, these data items were then defined by the attributes and their data types of the classes in vMR DAM [21]. The semantics of data items were further specified by reference to the guideline content. During the process of guideline modeling and data standardization, guideline deficiencies in clinical descriptions were revealed.
Second, we reviewed the data items and metadata defined in the national specification of patients' EHRs. By comparing them with those in the guideline ontology, conflicts and gaps were identified.
Finally, based on the previous steps, obstacles in computerizing guidelines were inferred and corresponding recommendations for the development of clinical guidelines and patient health record systems were made.
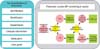
Based on clinical regulation, the hypertension guideline was disassembled into 6 parts: identification, risk stratification, classification of hypertension management, lifestyle intervention, medication, and care goals. These 6 parts and corresponding flowcharts are shown in Table 1. A full flowchart and overall 32 partitioned flowcharts, which are of various granularities, were created based on the rules described in the textual guideline. One of the flowcharts is shown in Figure 1, representing routine monitoring of blood pressure for adults.
All the rules in the flowcharts and corresponding statements represented in each node of the flowcharts are conceptualized in node and vMR. Flowchart, node, and vMR comprise the guideline ontology regarding medical knowledge of hypertension management.
In structuring and conceptualizing the guideline, one problem we encountered is inexplicit and imprecise narratives. Some statements, such as lack of physical exercise, long-term alcohol abuse, detect liver function when needed, increase examination frequency when disease worsens, etc., are qualitative and general; thus, they are very difficult to represent specifically in node of the ontology.
There are 140 data items extracted from node of the guideline ontology, which are classified into 4 classes in the ClinicalStatement package of vMR DAM: 17 data items to Goal, 45 to Observation, 13 to Problem, and 65 to SubstanceAdministration. Some are shown in Table 2.
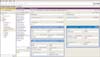
In defining all the data items, we also tried to localize the values of some attributes according to codes, identifiers, vocabularies, etc., provided in the target guideline. Figure 2 is a composite Protégé interface of browsing and editing classes and their instances. Taking the goal of blood pressure for general hypertension patients as an example, we defined the data items by specifying 2 attributes: goalFocus and targetGoalValue.
There were two major challenges in standardizing the data elements in localizing vMR DAM. The first challenge is how to tailor or specify the general definitions of vMR to satisfy the need to exactly represent the specific rules and concepts in the guideline. For example, targetGoalValue is defined with ANY, which is the most general and flexible HL7 data type. For the purpose of being more meaningful and computer processable, however, it would be better to have ANY be specified to PQ, QTY, BL, IVL, etc., that are capable of conveying explicit semantics, but such transformation is difficult without both clinical and informatics knowledge. Secondly, for the attributes whose values have been appointed to standard terminology or coding systems in vMR, such as SNOMED CT [22,23], they cannot be coded currently in China because of the unavailability of this standard. In China there is no alternative medical terminology system nationally. A similar example is LOINC, which has been widely used internationally [24], but it has not been widely recognized and adopted in China. So far, there have been neither mappings among various locally defined coding systems nor nationally unified identifiers for medical observations.
We reviewed the data items defined in the national specification of patients' EHRs of hypertension and analyzed the definitions provided in support documents. By comparing them with the standardized data definitions mentioned above, we found that among the data items extracted from the guideline, 8 items were left out in patients' records: age of family members when hypertension occurred, the cause of death of family members, measurement of urinary trace albumin and urinary albumin/creatinine, daily intake of sodium, and target for body weight control. Additionally, we found 10 data items that have definitions that are different from those of the guideline. Taking active health problems as an example, the guideline lists the diseases as cerebrovascular disease, heart disease, diabetes or impaired glucose tolerance, retinopathy, kidney disease, and peripheral vascular disease, while the patients' records categorize them as cerebrovascular, heart, neural, eye, kidney, and vascular diseases, as well as diseases of other systems. Another example is the kinship of family members. The guideline describes consanguinity as 1st, 2nd or 3rd generation, where patients' records include the relationships of father, mother, sibling and filial, which should all be grouped into 1st generation relationships.
The clinical guidelines provided by real-time CDSSs require that they are seamlessly integrated with existing patient information systems to enable the automatic provision of advice at the time and place at which decisions are made [25]. Focusing on hypertension, by modeling the knowledge and rules, our research first developed a guideline ontology to formulate the guideline and define the data items it contains. Secondly, we reviewed the health record specification for hypertension patients and compared the data definitions with those in the ontology to investigate whether they are mutually compatible in scope and semantics. Our investigation demonstrated that it is basically feasible to represent the guideline in a computer interpretable format. However, problems arose in defining semantics for some data items because of narrative ambiguity of the guideline, gaps and inconsistencies in representing some data items between the guideline and patient' records, and unavailability of unified medical coding systems. These problems might be common and need to be dealt with when any clinical guidelines are to be computerized. Accordingly, the following major suggestions for the development of guidelines and data specifications can be made.
Participation of people in different disciplines and at various levels is vital. Firstly, the development of a guideline requires input from, not only sufficiently qualified medical professionals who are able to make the right decisions in complex clinical situations, but also primary healthcare practitioners who need definitive directives when facing multiple choices. The participation of primary healthcare practitioners is even more important in the development of guidelines that target the management of long-term chronic diseases, such as hypertension, where general practitioners play leading roles. Therefore, the statements in a guideline should be more precise or knowledge intensive, making computerized CPGs and CDSSs more usable for people who really need them. Secondly, disease management specifications, which in China are designed mainly by public health agencies and address data recording issues, also need to be harmonious and consistent with the guidelines developed by clinical experts so as to facilitate guideline implementation. Obviously, this harmonization relies on effective communication and collaboration among all participants. Thirdly, difficulties in translating narrative guidelines to a computer interpretable format and in normalizing data definitions result partly from a lack of informaticians in the development of guideline and patients' record specifications. By identifying what kind of data standards or coding systems will be required in computerizing a specific guideline and whether they are available and applicable, informatics experts can certainly be helpful in solving some problems that have been identified in this research.
More attention should be paid to data standard development and adoption issues. Computable representation of clinical information requires lots of standards, especially medical terminologies so as to name, identify, and code medical concepts consistently. Unfortunately, few such standards have been developed domestically in China, and international standards have not been available so far. Therefore, we suggest that the government, healthcare, and health information communities make great effort to identify strategies, mechanisms, and technical solutions for clinical data standardization. Otherwise, it will be impossible to implement the guideline by CDSSs and to integrate data across various health information systems to build patient-centered, longitudinal EHR and clinical data repositories.
This study had several limitations. The guideline ontology built in this study is based on a guideline document modeled by our research team. It remains to be validated by clinical professionals in terms of its meaningfulness and accuracy. Additionally, this research investigated clinical data standardization issues only by focusing on the example of hypertension. It cannot be considered comprehensive, and there may be some other significant challenges remain unidentified. After all, integrating all the related data and knowledge in a CDSS has proven to be very complicated, while the variety of health information systems and clinical guidelines makes it even more difficult. Thus, the data standardization issue discussed herein is far from enough to execute a CDSS.
Figures and Tables
Figure 1
Flowchart for routine monitoring of BP in adults. BP: blood pressure, SBP: systolic BP, DBP: diastolic BP.
Acknowledgments
This research was supported by National High-Tech R&D Program (863 Program) of China (No. 2012AA02A603).
References
1. Isern D, Moreno A. Computer-based execution of clinical guidelines: a review. Int J Med Inform. 2008; 77(12):787–808.

2. Min YH, Park HA, Chung E, Lee H. Implementation of a next-generation electronic nursing records system based on detailed clinical models and integration of clinical practice guidelines. Healthc Inform Res. 2013; 19(4):301–306.

3. Eccher C, Purin B, Pisanelli DM, Battaglia M, Apolloni I, Forti S. Ontologies supporting continuity of care: the case of heart failure. Comput Biol Med. 2006; 36(7-8):789–801.

4. Mulyar N, van der Aalst WM, Peleg M. A pattern-based analysis of clinical computer-interpretable guideline modeling languages. J Am Med Inform Assoc. 2007; 14(6):781–787.

5. Peleg M, Tu S, Bury J, Ciccarese P, Fox J, Greenes RA, et al. Comparing computer-interpretable guideline models: a case-study approach. J Am Med Inform Assoc. 2003; 10(1):52–68.

6. de Clercq PA, Blom JA, Korsten HH, Hasman A. Approaches for creating computer-interpretable guidelines that facilitate decision support. Artif Intell Med. 2004; 31(1):1–27.

7. Peleg M, Keren S, Denekamp Y. Mapping computerized clinical guidelines to electronic medical records: knowledge-data ontological mapper (KDOM). J Biomed Inform. 2008; 41(1):180–201.

8. Tu SW, Campbell JR, Glasgow J, Nyman MA, McClure R, McClay J, et al. The SAGE Guideline Model: achievements and overview. J Am Med Inform Assoc. 2007; 14(5):589–598.

9. Johnson PD, Tu SW, Musen MA, Purves I. A virtual medical record for guideline-based decision support. Proc AMIA Symp. 2001; 294–298.
10. Liu L, Wang W, Yao C. [A guideline for prevention and medical care of hypertension in China (for primary care, 2009)]. Chin J Hypertens. 2010; 18(1):11–30.
11. Ministry of Health of People's Republic of China. China public health statistical yearbook 2010. Beijing, China: Peking Union Medical College Publishing House;2010.
12. Ministry of Health of People's Republic of China. Requirements of delivery of essential public health services [Internet]. Beijing, China: Ministry of Health of People's Republic of China;2012. cited at 2012 Dec 11. Available from: http://www.gov.cn/zwgk/2011-05/24/content_1870181.htm.
13. Cabana MD, Rand CS, Powe NR, Wu AW, Wilson MH, Abboud PA, et al. Why don't physicians follow clinical practice guidelines? A framework for improvement. JAMA. 1999; 282(15):1458–1465.

15. Tierney WM, Overhage JM, Takesue BY, Harris LE, Murray MD, Vargo DL, et al. Computerizing guidelines to improve care and patient outcomes: the example of heart failure. J Am Med Inform Assoc. 1995; 2(5):316–322.

16. Ahmadian L, van Engen-Verheul M, Bakhshi-Raiez F, Peek N, Cornet R, de Keizer NF. The role of standardized data and terminological systems in computerized clinical decision support systems: literature review and survey. Int J Med Inform. 2011; 80(2):81–93.

17. Chae YM, Yoo KB, Kim ES, Chae H. The adoption of electronic medical records and decision support systems in Korea. Healthc Inform Res. 2011; 17(3):172–177.

18. Real F, Riano D, Bohada J. Automatic generation of formal intervention plans based on the SDA representation model. In : Proceedings of the 20th IEEE International Symposium on Computer-Based Medical Systems; 2007 Jun 20-22; Maribor, Slovenia. p. 575–580.
19. Protégé wiki. Protégé user guide [Internet]. place unknown: Protégé wiki;c2014. cited at 2014 Mar 25. Available from: http://protegewiki.stanford.edu/wiki/Main_Page.
20. Chen CC, Chen K, Hsu CY, Li YC. Developing guideline-based decision support systems using Protégé and jess. Comput Methods Programs Biomed. 2011; 102(3):288–294.

21. HL7 Clinical Decision Support. Virtual medical record (vMR) for Clinical Decision Support: Domain Analysis Model, release 1 [Internet]. Ann Arbor (MI): Health Level Seven International;2011. cited at 2013 Jan 5. Available from: http://wiki.hl7.org/images/7/71/HL7vMR_vMR_Domain_Analysis_Model_Release_1.pdf.
22. International Health Terminology Standards Development Organization. SNOMED CT user guide (international release) [Internet]. Copenhagen, Denmark: International Health Terminology Standards Development Organization;c2013. cited at 2013 Sep 7. Available from: http://ihtsdo.org/fileadmin/user_upload/doc/download/doc_UserGuide_Current-en-US_INT_20130731.pdf.
23. Ahmadian L, Cornet R, de Keizer NF. Facilitating pre-operative assessment guidelines representation using SNOMED CT. J Biomed Inform. 2010; 43(6):883–890.

24. Regenstrief Institute. Logical Observation Identifiers Names and Codes (LOINC) users' guide [Internet]. Indianapolis (IN): Regenstrief Institute;c2013. cited at 2013 Sep 7. Available from: http://loinc.org/downloads/files/LOINCManual.pdf.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download