Abstract
Staphylococcus aureus is one of the major pathogens causing bovine mastitis and foodborne diseases associated with dairy products. To determine the genetic relationships between human and bovine or bovine isolates of S. aureus, various molecular methods have been used. Previously we developed an rpoB sequence typing (RSTing) method for molecular differentiation of S. aureus isolates and identification of RpoB-related antibiotic resistance. In this study, we performed spa typing and RSTing with 84 isolates from mastitic cows (22 farms, 72 cows, and 84 udders) and developed a molecular prophage typing (mPPTing) method for molecular epidemiological analysis of bovine mastitis. To compare the results, human isolates from patients (n = 14) and GenBank (n = 166) were used for real and in silico RSTing and mPPTing, respectively. Based on the results, RST10-2 and RST4-1 were the most common rpoB sequence types (RSTs) in cows and humans, respectively, and most isolates from cows and humans clearly differed. Antibiotic resistance-related RSTs were not detected in the cow isolates. A single dominant prophage type and gradual evolution through prophage acquisition were apparent in most of the tested farms. Thus, RSTing and mPPTing are informative, simple, and economic methods for molecular epidemiological analysis of S. aureus infections.
Staphylococcus aureus causes bovine mastitis and food poisoning and is an important pathogen in the dairy industry [2540]. Since 1961, methicillin-resistant S. aureus (MRSA) has become an important bacterial threat worldwide. Molecular epidemiological investigations have been performed using various isolates of S. aureus from humans and animals [1542]. Although comparative genomics has been successfully applied to elucidate the transmission and evolution of whole genetic contents related to antibiotic resistance and pathogenicity, it remains costly and time-consuming when analyzing large datasets. Sub-genomic methods including multi-locus sequence typing (MLST), arbitrary primer-polymerase chain reaction (PCR), pulsed-field gel electrophoresis, multi-locus variable number tandem repeat analysis, spa typing, small genomic islet typing, rpoB sequence typing (RSTing), and microarray hybridization have been used [58112627283341]. Among these methods, RSTing has been developed to reduce the experimental cost of MLST and was successfully applied to the molecular epidemiological study of human and avian isolates of S. aureus [33]. Furthermore, RSTing provides information on rifampin, daptomycin, and vancomycin resistance in S. aureus, which are useful for MRSA treatment [164546].
Prophage typing after induction with mitomycin C has been used to characterize S. aureus strains [21]. Temperate bacteriophages (phages) carry and mobilize diverse virulence genes, and prophage profiles may help predict the potential pathogenicity of S. aureus isolates [214202944]. The terminase large subunit gene was used to identify prophages in bacterial genomes and for the differentiation of phages from environments and uncultured bacteria [3435]. A molecular prophage typing (mPPTing) method based on the terminase large subunit gene has been developed for molecular epidemiological study of avian pathogenic Escherichia coli in chickens and has provided additional information regarding phage-carrying virulence genes [24]. In addition, mPPTing with integrase genes in temperate phages of S. aureus has been developed [9].
In the present study, we developed a novel mPPTing method by using terminase large subunit genes for 84 and 14 isolates of S. aureus from bovine mastitis and humans, respectively, and compared the results with those from in silico mPPTing of 166 human S. aureus strains in the GenBank database. This mPPTing method provides information about prophage profiles carrying virulence genes as well as on the molecular epidemiology of S. aureus infection.
A total of 84 S. aureus isolates from bovine mastitis cows (22 farms, 72 cows, and 84 udders) in Korea were isolated after cultivation on sheep blood agar and identified by using the VITEK2 system and Gram-Positive Identification Cards (BioMérieux, France) [18]. Fourteen human MRSA isolates were purchased from the Culture Collection of Antimicrobial Resistant Microbes (Korea), and MLST and rpoB sequence type (RST) were characterized previously [33] (Supplementary Table 1). The 166 human strains were collected from the genome sequences of S. aureus in the GenBank database (Supplementary Table 1).
The primer sets for RSTing and detection of the methicillin-resistance gene, mecA, and spa typing were used as previously reported [193338]. The primer sets for mPPTing were designed by using large subunit terminase genes of S. aureus collected from the GenBank database; primer sequences are summarized in Table 1. Primer sets for detecting prophage-related virulence genes, as well as staphylokinase (sak) and chemotaxis-inhibiting (cip) genes, were designed and used in this study (sakF, 5′-GCGAT GACGC GAGTT ATTTT-3′ [116–135]; sakR, 5′-GACTT CGATC TTTGC GCTTG-3′ [329–348]; cipF, 5′-GCAGG AATCA GTACA CACCA TCATT CAG-3′ [49–76]; cipR, 5′-CAGCA AGTGG TGTAT TCAGA TATAC TGTAT AG-3′ [297–328]).
The S. aureus strains were grown overnight in tryptic soy broth at 37℃, and genomic DNA was extracted from 1 mL of S. aureus tryptic soy broth culture by using a G-spin For Bacteria Genomic DNA Extraction Kit (iNtRON Biotechnology, Korea) according to the manufacturer's instructions. The rpoB PCR mixture was composed of 5 µL of 10× reaction buffer, dNTPs (10 mM, 2.5 µL), Exprime Taq DNA polymerase (5 U/µL; Genet Bio, Korea; 0.5 µL), forward and reverse primers (1. 25 µM, 1.25 µL of each), distilled water (38.5 µL), and template DNA (50 ng/µL, 1 µL). The mixture was incubated at 94℃ for 5 min; 35 cycles of 94℃ for 30 sec–54℃ for 30 sec–72℃ for 4 min with a final extension step at 72℃ for 5 min. The PCR for spa typing was performed as described for rpoB PCR except for the PCR condition: 80℃ for 5 min; 35 cycles at 94℃ for 45 sec–60℃ for 45 sec–72℃ for 90 sec with a final extension step at 72℃ for 10 min [38]. The PCR for mPPTing and for sak and cip were performed as described for rpoB PCR except for elongation times of 2 min and 30 sec, respectively, were used for the 35 cycles. PCR for mecA was performed as described previously [19].
The PCR amplicons were purified using the MEGAquick-spin Total Fragment DNA Purification Kit (iNtRON Biotechnology) and sequenced with sequencing primers using an ABI3711 automatic sequencer (Macrogen, Korea).
Overlapping sequences of the rpoB genes of S. aureus isolates from bovine mastitis were assembled into single complete sequences by using ChromasPro (ver. 1.5; Technelysium, Australia). After comparison with consensus and previously identified RST sequences, the RST of the new rpoB sequence was assigned as previously described [33]. Nucleotide similarity, variable nucleotide comparisons, and translation of nucleotide sequences were performed with Bioedit software (ver. 5.0.9.1; Ibis Biosciences, USA). Phylogenetic analyses with complete rpoB genes were conducted using MEGA software (ver. 7; neighbor-joining method with p-distance and 1,000 repeats of bootstrapping) [23]. The spa type of each S. aureus isolate was determined by using BioNumerics (ver. 7.6; Applied Math, Belgium).
In silico mPPTing was performed by querying the representative sequence of each prophage group by BLAST searching and confirming 100% nucleotide matches of the primer regions. In silico MLST was performed by querying the retrieved housekeeping genes of reference strains in which the sequence type (ST) was not described in the GenBank files by using BLAST based on the allele templates in the MLST database.
The index of diversity (ID) and confidence interval (CI, 95%) were calculated as previously described [10]. The frequencies of RSTs, spa types, phage groups, and molecular prophage types (mPPTs) were compared via chi-squared and Fisher's exact tests (95% CI) using SPSS for Windows (ver. 12.0; SPSS, USA).
The complete rpoB coding region of S. aureus composed of 3,552 nucleotides was successfully amplified by the PCR primer set. The complete rpoB sequences of the bovine S. aureus isolates in this study were deposited in GenBank (MG737590–MG737673) and the accession numbers of all rpoB sequences used for analysis are separately shown in Fig. 1 and Supplementary Table 1. Nucleotide similarities between rpoB sequences were 99.0% to 100%, and the amino acid changes from the consensus sequence are summarized in Table 2. Amino acid changes related to rifampin, daptomycin, and vancomycin resistance are also summarized in Table 2. No bovine S. aureus isolates showed amino acid changes related to rifampin, daptomycin, and vancomycin resistance [14546]. Therefore, they may have no or low resistance to those antibiotics.
The complete rpoB sequences of 84 bovine isolates were compared with the consensus sequence and classified into 13 RSTs as previously described [33] (Table 3). We collected all rpoB sequences from human, animals, and the environment in the GenBank database and classified these sequences into RSTs. A total of 89 RSTs from RST2-1 to RST25-1 were assigned to all S. aureus isolates and strains (Table 4). The representative rpoB sequence of each RST was used for phylogenetic analysis and these sequences formed distinct clusters with each other in the phylogenetic tree (Fig. 1). Particularly, RSTs with neighboring numbers, such as 2–3, 4–7, 8–11, 14–17, and 21–23, formed a distinct cluster with other RSTs. The frequencies of RSTs of bovine isolates and human strains of S. aureus are summarized in Table 3. RSTs 10-2 (41.7%) and 4-1 (37.1%) were the most common among bovine isolates and human strains, respectively, and accounted for 51.2% and 48.9% of isolates, respectively, to sum up the first and the second most predominant genotypes of bovine isolates (RST10-3) and human strains (RST2-1). The RSTs 2-1, 4-1, 5-2, 8-1, 10-2, 14-2, and 14-3 were commonly present in bovine isolates and human strains, while RSTs 10-3, 11-4, 11-5, 11-6, 11-7, and 22-1 were only detected in bovine isolates. The frequency of RST10-2 in bovine isolates (41.7%, 35/84) was significantly higher than that (2.8%, 5/180) in human isolates of S. aureus (p < 0.05).
The discriminative power of RSTing and MLST was compared with those of actual and in silico results presented in Supplementary Table 1. The RSTs 2-1, 4-1, 7-1, 10-1, 10-2, and 14-2 were composed of 4, 8, 2, 2, 2, and 2 different STs, respectively. The 5, 8, 239, 72, 22, 1, 398, and 45 STs were composed of 6, 5, 6, 2, 2, 2, 5, and 2 different RSTs. Thus, RSTing could not differentiate 20 STs, while MLST could not differentiate 30 RSTs. However, the IDs (CIs) of RSTing and MLST were 85.1% (80.6–89.5%) and 86.7% (82.6–90.8%), respectively, when they were calculated on the basis of data in Supplementary Table 1, except for the not tested and untypable (UT) strains. Therefore, RSTing showed slightly less discriminative power than MLST.
The spa types of 84 bovine isolates were determined (Table 3). Eighty isolates were assigned into 10 spa types, t127 (26.2%, 22/84), t189 (19.0%, 16/84), t002 (15.5%, 13/84), t084 (8.3%, 7/84), t034 (7.1%, 6/84), t304 (7.1%, 6/84), t164 (6.0%, 5/84), t2459 (2.4%, 2/84), t4050 (2.4%, 2/84), t2612 (1.2%, 1/84). Four isolates were classified into 4 UT variants, PMB 173-1 (11-10-21-17-172-24-34-22-25, UT1), PMB 188-1 (11-19-17-34-22-25-25-25, UT2), PMB 8-1 (26-17-34-34-34-34-33-34, UT3), PMB 8-2 (26-17-34-34-34-34-22-34-34-34-34-33-34, UT4). Most of the isolates from the same farm showed the same spa type and the farm-based frequency of each spa type was as follows: 13.6% (3/22; t127, t189, and t002), 9.1% (2/22; t034, t164, and t4050), and 4.5% (1/22; t084, t304, t2612, t2459, UT1, UT2, UT3, and UT4). The major RST10-2 was composed of 4 spa types including two major spa types, t127 and t189. Relatively high frequencies of t127 and t189 among S. aureus isolates of bovine mastitis have already been reported in Korea and Brazil, and South Africa, respectively [133236].
The terminase large subunit genes of prophages were collected from the GenBank database by keyword and BLAST searches and then subjected to phylogenetic analysis. Terminase genes forming a cluster were aligned and a cluster-specific PCR primer set was designed (Table 1). The PCR results are shown in Fig. 2. Arabic numbers were assigned to prophage groups (1, 2, 3, 5, 6, 7, 8, 9, 10, and 12), and a representative phage from each prophage group with high nucleotide identity (not less than 99%) is summarized with its name, accession number, and genome size in Table 1. All representative phages were classified into Family Siphoviridae, and φNM2, φ13, φSPβ-like, and φETA3 have been reported to possess virulence genes [21420]. Eighty-four and 14 cow isolates and human strains, respectively, were tested for the presence of each prophage group. Prophage group 1 (40.5%) was the most frequent in bovine isolates, followed by phage groups 7 (26.2%), 2 (23.8%), 3 (21.4%), 10 (17.9%), 8 (11.9%), and 9 (11.9%). Phage groups 5, 6, and 12 were not detected. The frequency of phage groups of S. aureus isolates from human patients were combined with in silico results that were predicted by using genome sequences of human S. aureus strains in the GenBank database. According to the results, phage groups 7 and 9 (69.4%) were the most frequent, followed by phage groups 1 (55.1%), 2 (25.7%), 3 (13.8%), 5 (10.2%), 8 (7.2%), 10 (6.6%), 12 (4.2%), and 6 (3.0%). The frequencies of phage groups 7 and 9 in human isolates were significantly higher than those in bovine isolates of S. aureus (p < 0.05).
The real and in silico mPPTs of each S. aureus isolate are summarized in Table 4 and Supplementary Table 1. The mPPT is represented as a hyphen-separated number of prophage groups possessed by a given isolate with mPPT0 assigned to isolates without a detected prophage group. Each RST was divided into subgroups of mPPTs, and S. aureus isolates from the same farm or the same individual showed the same or similar mPPTs. Interestingly, the genotype, RST10-2/mPPT0 was dominant compared to other genotypes and was distributed on a limited number of dairy farms (4/26 farms, 15.4%). The frequency of mPPT0 among different RSTs was higher in bovine isolates (6/13 RSTs, 46.2%) than in human strains (10/49 RSTs, 20.4%), but the difference was not significant. In addition, mPPT7-9 (31/180, 17.2%) was the most frequent in human strains, followed by mPPTs 1-7-9 (30/180, 16.7%) and 1-2-7-9 (30/180, 16.7%). Step-wise, cumulative acquisition of the prophage was apparent in RST10-2 and RST11-4, and RST2-1 and RST4-1 of bovine isolates and human strains, respectively. The mPPTs possessing at least 5 prophage groups were absent in bovine isolates but present in the 2-1, 4-1, 5-1, 6-2, 6-4, and 6-8 RSTs of human strains.
Bovine isolates with the same RSTs/mPPTs as human strains were present. RST10-2/mPPT0, RST10-2/mPPT1-3, RST14-2/mPPT1-7-9, and RST5-2/mPPT7-9 were also present in human strains, and multiple strains of RST10-2/mPPT1-3 and RST5-2/mPPT7-9 were present in 5 and 3 strains, respectively.
We calculated IDs and CIs of spa typing, RSTing, and mPPTing for 84 bovine isolates of S. aureus. The IDs (Cis) of spa typing, RSTing, and mPPTing were 85.9% (82.2–89.6%), 79.1% (71.7–86.5%), and 90.3% (87.8–92.8%), respectively. Thus, spa typing had higher discriminative power than RSTing but less than that of mPPTing.
In most cases, S. aureus isolates from a farm had the same RSTs and the same or slightly different mPPTs. Of the 13 multi-sampled dairy farms, a single genotype of S. aureus was observed on 5 farms (PMB8, PMB119, PMB177, PMB179, and PMB238; 38.5%) and mixed genotypes composed of parent and progeny genotypes of S. aureus were observed on 7 farms (PMB61, PMB81, PMB132, PMB196, PMB208, PMB232, and PMB242; 53.8%). Evolution of the dominant genotype via acquiring new prophages on dairy farms of mixed genotypes was also apparent (Fig. 3). The mPPT10 (PMB61-1-1, PMB61-1-2, and PMB242-4) and mPPT7 (PMB196-1, 2, 3, 5-2, and 6) acquired a prophage group 8 phage to become mPPT8-10 (PMB61-1-3, PMB242-1, 2, and 3) and mPPT7-8 (PMB196-4, 5-1, 7, and 8), respectively. The mPPT0 (PMB231-2; PMB132-1, 2, 3, 4, 6, and 7; PMB208-3) became mPPT1 (PMB232-1 and PMB132-5) and mPPT7-9 (PMB208-1 and 2) by acquiring prophage group 1 and 7-9 phages, respectively. In addition, mPPT1-2 (PMB81-11) and mPPT1-3 (PMB81-8) became mPPT1-2-3 by acquiring prophage groups 2 and 3 phages, respectively. Therefore, transmission of the dominant genotype of S. aureus between individual cows and evolution by the acquisition of prophages on dairy farms was commonly observed.
MecA was not detected in any isolate and might be infrequent in Korean bovine isolates. The representative phages of prophage groups 3 and 7 are φ13 and φP282, respectively. Phages φ13 (sak) and φP282 (sak and cip) carry virulence genes; thus, we tested the presence of the virulence genes in the bovine isolates. Of prophage group 3-positive isolates (PMB81-1 and PMB232-1), PMB232-1 possessed sak, and of prophage group 7-positive isolates (PMB67-1, PMB177-1, PMB188-1, PMB196-1, and PMB208-1), PMB67-1, PMB196-1, and PMB208-1 contained sak and cip (Fig. 4).
Various sub-genomic molecular typing methods have been successfully applied to S. aureus isolates, and such methods can provide information regarding genetic diversity for use in molecular epidemiological studies. Among these methods, MLST has been widely applied to various pathogenic bacteria, but its handling of multiple genes, low discriminative power at the serotype level, and lack of clinical information for the data limit the applicability of this method. Thus, we developed the RSTing method for molecular serotyping of Salmonella enterica subsp. enterica and examining the molecular epidemiology of S. aureus isolates [3334].
In this study, we used RSTing to investigate the molecular epidemiology of bovine mastitis caused by S. aureus isolates. The predominance of certain genotypes of S. aureus in different host species has been reported, which was confirmed by the results of the RSTing method [7173339]. RSTs 10-2 and 10-3, and 4-1, and 2-1 were predominant, accounting for approximately 50% of bovine isolates and human strains. The greater diversity among the RSTs of human strains (48) compared to that of bovine isolates (13) supports the observation that S. aureus of humans spread to animals and has adapted to specific niches in animals [3031]. The presence of RSTs 2-1 and 4-1 in bovine isolates may reflect human to animal transmission, but transmission by contact with humans may occur occasionally based on the relatively low frequencies of the RSTs. However, transmissions of animal-specific S. aureus to humans was commonly observed and livestock-associated MRSA (LA-MRSA) has been reported [1243]. RST10-2 was observed in human strains, and FORC 026 and FORC 045 strains isolated from Korean patients completely matched the Korean bovine isolates' mPPTs. These strains may be transmitted occasionally by foodborne diseases. Therefore, recent, not infrequent transmission of LA-MRSA from pigs and cows to humans may be a reason for the application of RSTing and mPPTing methods to unravel epidemiological relatedness between them [1237].
The first RST identifier number represents the number of nucleotides mismatched to the consensus sequence, and the clustering of RSTs 2–3, 4–7, 8–11, 14–17, or 20–23 may reflect phylogenetic relationships among these sequences. Therefore, the first RST identifier number reflects the phylogenetic relationships with other RSTs. Furthermore, a comparable discriminative power, but simpler and more economic protocol of RSTing than that of MLST, may indicate the merits of using RSTing [33]. Although the discriminative power of RSTing was less than spa typing the antibiotic resistance information from RSTing data may support it being the preferred choice as a frontline tool. Additionally, the higher discriminative power of mPPTing than spa typing may also support it being the preferred choice as a second-line tool when combined with RSTing of molecular epidemiology of S. aureus infections in animals and humans.
The evolution of S. aureus is largely driven by mobile genetic materials such as temperate phages, plasmids, transposons, and mobile pathogenicity islands [29]. Among them, temperate phages have key roles in the transmission and expression of virulence genes of S. aureus, and investigations of the prophage profiles of S. aureus isolates may be useful in elucidating the molecular evolution and epidemiology of S. aureus. Most virulence-related temperate phages possess integrases and are classified into Family Siphoviridae [9]. Because integrases are not shared by lytic phages, we chose to investigate terminase large subunit genes common to both lysogenic and lytic phages [3435].
The different frequencies of prophage groups 7 and 9 between bovine isolates and human strains may reflect different kinetics among temperate phages. The absence of prophage groups 5, 6, and 12 and more RSTs without prophages (mPPT0) in bovine isolates than in human strains may reflect a lower probability of phage transduction and less diverse temperate phage pools in bovine isolates. However, the presence of mPPTs possessing at least 5 prophage groups may also reflect frequent phage transduction and diverse temperate phage pools in human S. aureus strains.
Among the multi-sampled dairy farms, the frequency of mixed genotypes composed of parent and progeny genotypes of S. aureus was more than half (53.8%), and the evolution of bovine isolates by the acquisition of prophages on dairy farms was evident. Additionally, the presence of a single genotype among mastitic cows on a farm should encourage efforts to prevent horizontal transmission. The relatively high frequency of prototypic RST10-2/mPPT0 on different farms (4/26, 15.4%) reflects the method of transmission and introduction onto farms, as well as its role as a founder in the evolution of various mPPTs.
Although LA-MRSA has been reported worldwide, the absence of mecA and antibiotics-resistance-related RSTs among bovine isolates in this study reflects a minor potential threat of bovine S. aureus transmission to humans [162243]. The presence of virulence genes (sak and cip) in the mPPT 3 and 7 bovine isolates may be useful for predicting the pathotypes of S. aureus isolates.
In this study, we applied RSTing to elucidate the molecular epidemiology of bovine mastitis caused by S. aureus and developed mPPTing to evaluate the evolution of RSTs. According to the results, bovine- and human-specific predominant RSTs of S. aureus were identified, and a gradual evolution of S. aureus RSTs, via prophage acquisition, was evident on the dairy farms. Thus, a combination of RSTing and mPPTing may be an economic and informative method for understanding the molecular epidemiology of animal and human diseases caused by S. aureus infection.
Figures and Tables
Fig. 1
Phylogenetic analysis of the complete rpoB sequences of 90 representative rpoB sequence types and consensus sequence of Staphylococcus aureus. The phylogenetic tree was constructed by using the neighbor-joining method (p-distance and 1,000 bootstrapping replicates) with MEGA software (ver. 7) [23]. The bovine isolates analyzed in this study are marked with closed circles.
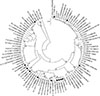
Fig. 2
Amplification of terminase large subunit genes of prophage groups 1, 2, 3, 6, 7, 8, 9, and 10. Amplicons of prophage groups 5 and 12 are not shown because positive bovine isolates and human strains were not available. Lane M, 1000 bp size marker; Lane 1, prophage group 1 (1585 bp); Lane 2, prophage group 2 (1212 bp); Lane 3, prophage group 3 (813 bp); Lane 4, prophage group 6 (575 bp); Lane 5, prophage group 7 (895 bp); Lane 6, prophage group 8 (1116 bp); Lane 7, prophage group 9 (1223 bp); Lane 8, prophage group 10 (1769 bp).
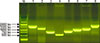
Fig. 3
Evolution of Staphylococcus aureus on dairy farms through the acquisition of temperate phages. The first, second, and third numbers of each isolate's name are identifiers of the farm, individual cow, and udder, respectively. mPPT, molecular prophage type; + pp, acquisition of prophage.

Fig. 4
Virulence genes of prophage groups 3 and 7. The staphylokinase (A) and chemotaxis inhibiting (B) genes were detected by polymerase chain reaction. (A) Lane M, 100 bp size marker; Lane 1, PMB 81-1; Lane 2, PMB 232-1; Lane 3, PMB 67-1; Lane 4, PMB 177-1; Lane 5, PMB 188-1; Lane 6, PMB 196-1; Lane 7, PMB 208-1. (B) Lane M, 100 bp size marker; Lane 1, PMB 67-1; Lane 2, PMB 177-1; Lane 3, PMB 188-1; Lane 4, PMB 196-1; Lane 5, PMB 208-1.

Acknowledgments
This research was supported by a Research Program through the Rural Development Administration (RDA) funded by the Ministry of Agriculture, Food and Rural Affairs (grant No. PJ010855). We appreciate Prof. Yong-Ho Park and Ms. Sook Shin (Department of Microbiology, College of Veterinary Medicine, Seoul National University) for providing S. aureus strains (PMB66, PMB67, PMB132, and PMB146).
References
1. Aubry-Damon H, Soussy CJ, Courvalin P. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1998; 42:2590–2594.

2. Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol. 2006; 62:1035–1047.

4. Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003; 49:277–300.

5. Cassat JE, Dunman PM, McAleese F, Murphy E, Projan SJ, Smeltzer MS. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J Bacteriol. 2005; 187:576–592.

6. Cui L, Isii T, Fukuda M, Ochiai T, Neoh HM, Camargo IL, Watanabe Y, Shoji M, Hishinuma T, Hiramatsu K. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2010; 54:5222–5233.

7. Devriese LA, Oeding P. Characteristics of Staphylococcus aureus strains isolated from different animal species. Res Vet Sci. 1976; 21:284–291.

8. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000; 38:1008–1015.

9. Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Bröker BM, Doskar J, Wolz C. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol. 2009; 191:3462–3468.

10. Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. 2001; 39:4190–4192.

11. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003; 41:5442–5448.

12. Huijsdens XW, van Dijke BJ, Spalburg E, van Santen-Verheuvel MG, Heck ME, Pluister GN, Voss A, Wannet WJ, de Neeling AJ. Community-acquired MRSA and pig-farming. Ann Clin Microbiol Antimicrob. 2006; 5:26.
13. Hwang SY, Park YK, Koo HC, Park YH. spa typing and enterotoxin gene profile of Staphylococcus aureus isolated from bovine raw milk in Korea. J Vet Sci. 2010; 11:125–131.

14. Iandolo JJ, Worrell V, Groicher KH, Qian Y, Tian R, Kenton S, Dorman A, Ji H, Lin S, Loh P, Qi S, Zhu H, Roe BA. Comparative analysis of the genomes of the temperate bacteriophages φ11, φ12 and φ13 of Staphylococcus aureus 8325. Gene. 2002; 289:109–118.

16. Juhász-Kaszanyitzky E, Jánosi S, Somogyi P, Dán A, van der Graaf-van Bloois L, van Duijkeren E, Wagenaar JA. MRSA transmission between cows and humans. Emerg Infect Dis. 2007; 13:630–632.

17. Kapur V, Sischo WM, Greer RS, Whittam TS, Musser JM. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J Clin Microbiol. 1995; 33:376–380.

18. Kim D, Kim EK, Seong WJ, Ro Y, Ko DS, Kim NH, Kim JH, Kwon HJ. Identification of microbiome with 16S rRNA gene pyrosequencing and antimicrobial effect of egg white in bovine mastitis. Korean J Vet Res. 2017; 57:117–126. Korean.

19. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007; 51:264–274.

20. Kraushaar B, Hammerl JA, Kienöl M, Heinig ML, Sperling N, Dinh Thanh M, Reetz J, Jäckel C, Fetsch A, Hertwig S. Acquisition of virulence factors in livestock-associated MRSA: lysogenic conversion of CC398 strains by virulence gene-containing phages. Sci Rep. 2017; 7:2004.

21. Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983; 305:709–712.

22. Kuehn BM. Antibiotic-resistant "superbugs" may be transmitted from animals to humans. JAMA. 2007; 298:2125–2126.

23. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1874.

24. Kwon HJ, Seong WJ, Kim JH. Molecular prophage typing of avian pathogenic Escherichia coli. Vet Microbiol. 2013; 162:785–792.
25. Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genet Mol Res. 2003; 2:63–76.
26. Lee JH. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. 2003; 69:6489–6494.

27. Malachowa N, Sabat A, Gniadkowski M, Krzyszton-Russjan J, Empel J, Miedzobrodzki J, Kosowska-Shick K, Appelbaum PC, Hryniewicz W. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J Clin Microbiol. 2005; 43:3095–3100.

28. Monecke S, Ruppelt A, Wendlandt S, Schwarz S, Slickers P, Ehricht R, Jäckel SC. Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet Microbiol. 2013; 162:806–812.
29. Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003; 49:93–105.

30. Resch G, François P, Morisset D, Stojanov M, Bonetti EJ, Schrenzel J, Sakwinska O, Moreillon P. Human-to-bovine jump of Staphylococcus aureus CC8 is associated with the loss of a β-hemolysin converting prophage and the acquisition of a new staphylococcal cassette chromosome. PLoS One. 2013; 8:e58187.
31. Sakwinska O, Giddey M, Moreillon M, Morisset D, Waldvogel A, Moreillon P. Staphylococcus aureus host range and human-bovine host shift. Appl Environ Microbiol. 2011; 77:5908–5915.

32. Schmidt T, Kock MM, Ehlers MM. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: genetic diversity and inter-species host transmission. Front Microbiol. 2017; 8:511.

33. Seong WJ, Kim JH, Kwon HJ. Comparison of complete rpoB gene sequence typing and multi-locus sequence typing for phylogenetic analysis of Staphylococcus aureus. J Gen Appl Microbiol. 2013; 59:335–343.

34. Seong WJ, Kwon HJ, Kim TE, Lee DY, Park MS, Kim JH. Molecular serotyping of Salmonella enterica by complete rpoB gene sequencing. J Microbiol. 2012; 50:962–969.

35. Serwer P, Hayes SJ, Zaman S, Lieman K, Rolando M, Hardies SC. Improved isolation of undersampled bacteriophages: finding of distant terminase genes. Virology. 2004; 329:412–424.

36. Silva NC, Guimarães FF, Manzi MP, Budri PE, Gómez-Sanz E, Benito D, Langoni H, Rall VL, Torres C. Molecular characterization and clonal diversity of methicillin-susceptible Staphylococcus aureus in milk of cows with mastitis in Brazil. J Dairy Sci. 2013; 96:6856–6862.

37. Spoor LE, McAdam PR, Weinert LA, Rambaut A, Hasman H, Aarestrup FM, Kearns AM, Larsen AR, Skov RL, Fitzgerald JR. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. MBio. 2013; 4:e00356–e00313.

38. Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nübel U, Witte W. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008; 46:574–581.

39. Sung JM, Lloyd DH, Lindsay JA. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology. 2008; 154:1949–1959.

40. Sutra L, Poutrel B. Virulence factors involved in the pathogenesis of bovine intramammary infections due to Staphylococcus aureus. J Med Microbiol. 1994; 40:79–89.

41. Suzuki M, Matsumoto M, Takahashi M, Hayakawa Y, Minagawa H. Identification of the clonal complexes of Staphylococcus aureus strains by determination of the conservation patterns of small genomic islets. J Appl Microbiol. 2009; 107:1367–1374.

42. Thompson RL, Cabezudo I, Wenzel RP. Epidemiology of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. Ann Intern Med. 1982; 97:309–317.

43. van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007; 13:1834–1839.

44. van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J Bacteriol. 2006; 188:1310–1315.





 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download