Abstract
A novel porcine circovirus 3 (PCV3) was first detected in pigs showing porcine dermatitis and nephropathy syndrome, reproductive failure, and multisystemic inflammation in the USA. Herein, we report on PCV3 as a potential etiological agent of clinical signs, reproductive failure and respiratory distress on Korean pig farms, based on in situ hybridization, pathological, and molecular findings. Confirmation of the presence of PCV3 may increase co-infection with other causative agents of disease in Korean pig herds, indicating the need for further systemic investigation of pathogenicity and of multiple infections with PCV2 genotypes and bacteria, and the development of an effective PCV3 vaccine.
Circoviruses are non-enveloped, spherical viruses with a small monomeric single-stranded circular DNA genome of 1,759 to 1,768 nucleotides [5]. Two genotypes of porcine circovirus (PCV), PCV1 and PCV2, reportedly infect pigs. While PCV1 is considered non pathogenic [18], PCV2 is a major pathogen of porcine circovirus disease, which is characterized by reproductive disorders, enteritis, postweaning multisystemic wasting syndrome (PMWS), porcine dermatitis and nephropathy syndrome (PDNS), proliferative and necrotizing pneumonia, and porcine respiratory disease complex [13]. Five PCV2 genotypes are recognized: PCV2a, 2b, 2c, 2d, and 2e [4]. The genetic diversity of PCV2 strains has continuously increased, and co-existence of different PCV2 genotypes in one pig herd has been reported [819]. Palinski et al. [11] identified a highly divergent new genotype of PCV, designated PCV3, from sows with PDNS-like clinical signs and mummified fetuses of various gestational ages. Furthermore, Phan et al. [12] reported the detection of PCV3 in wasted pigs with cardiac pathology and multisystem inflammation at 2 to 3 and 9 to 10 weeks of age. Porcine astrovirus was also detected in those animals. In this study, we report the first detection and molecular characterization of PCV3 from pigs with reproductive failure and respiratory distress on Korean domestic pig farms. The full-length genome sequences of two strains were determined and compared with those of Korean PCVs and recently reported strains in other countries, including the USA and China.
Between November 2016 and January 2017, a fattening pig farm with an inventory of 1,200 sows located in North Gyeongsang Province, in the southeast of Korea, experienced a 10% increase in the sow abortion rate and a 20% increase in the suckling pig death rate above the historical average abortion rate. A veterinarian submitted autopsy samples of 14 aborted fetuses of gestational age 60 to 100 days from 6 sows and 8 weak 3- to 7-day-old suckling pigs from 4 sows to the Animal Disease Intervention Center at Kyungpook National University, Korea for etiological diagnosis. Total nucleic acids were extracted from the supernatant of each submitted sample (lungs and hearts of aborted fetuses and lungs, hearts, lymph nodes, kidneys, spleens, and livers of suckling pigs) and subjected to polymerase chain reaction (PCR) and reverse transcriptase (RT)-PCR using commercial kits (Median, Korea) to determine the presence of known abortifacient viruses. For detection of Brucella spp., Leptospira spp., and Toxoplasma gondii, the tissue samples were subjected to PCR or real-time PCR [1416]. Published primers and probes were used for PCV3 detection and sequencing [11]. All tested tissue samples were negative for porcine reproductive and respiratory syndrome virus (PRRSV), PCV2, porcine parvovirus 1 (PPV1), encephalomyocarditis virus, Aujeszky's disease virus, classical swine fever virus (CSFV), Japanese encephalitis virus, swine influenza virus (SIV), and for Brucella spp., Leptospira spp., and T. gondii based on PCR analysis. By contrast, PCV3 was detected in 6 (quantification cycle [Cq] value, 27–30) of 14 aborted fetuses and 2 (Cq value, 21–30) of 8 suckling pig samples by real-time PCR. The PCV3 positive rates were highest in lungs of aborted fetuses and kidneys from suckling pigs (Table 1).
In mid-October 2016, a farrow-to-finish farm with an inventory of 150 sows located in South Gyeongsang Province in the southeast of Korea experienced wasting and respiratory distress in 2% of growing pigs, and appropriate samples were submitted to the Animal Disease Diagnostic Division of the Animal and Plant Quarantine Agency. Two dead pigs were subjected to postmortem examinations, and lung, liver, kidney, heart, spleen, tonsil, and lymph node samples were collected. The samples were analyzed by RT-PCR for PRRSV, CSFV, and SIV, as well as by PCR for PCV2 and PPV1, by using commercial diagnostic kits (Median). The structural gene of PCV3 was PCR-amplified using PCV3 specific primers and probes [11]. Direct detection of Mycoplasma hyorhinis and Mycoplasma hyopneumoniae was performed by PCR using primers reported elsewhere [1]. Lung samples were cultured on 5% sheep blood agar and chocolate agar (Asan Pharm, Korea) under 5% CO2 at 37℃ for 24 h. The major internal organs were fixed in 10% phosphate-buffered formalin and routinely processed. The processed tissues were embedded in paraffin and stained with hematoxylin and eosin for light microscopy examination. To detect PCV3 in the lungs and lymph nodes of infected pigs, in situ hybridization (ISH) was performed using a PCV3-specific probe generated by using a PCR DIG Probe Synthesis Kit (Roche Diagnostics, Germany). The ISH was performed by using a fully automated system and a DAB detection system (NexES IHC instrument; Ventana Medical Systems, USA).
No PCV2, CSFV, or SIV nucleic acid was detected by PCR or RT-PCR in any of the tested samples; however, PRRSV-1 RNA and PCV3 DNA (Cq value, 17–21) were detected in both specimens by RT-PCR and real-time PCR, respectively. No bacteria were isolated or detected; however, M. hyopneumoniae was detected by PCR. Streptococcus suis and Haemophilus parasuis were examined by PCR [6] based on confirmation of polyserositis in the internal organs. Finally, S. suis was detected as a co-infecting pathogen with M. hyopneumoniae in the lung. Gross lesions in the lungs were characterized by a lack of pulmonary collapse with moderate-to-severe interstitial edema. Cranioventral, lobular to lobar bronchopneumonia was also observed. The submandibular and tracheobronchial lymph nodes showed severe enlargement. Furthermore, yellowish fibrinous materials were attached to the serosa of the heart and liver. Histopathologically, the lungs showed focal to diffuse bronchointerstitial pneumonia with macrophages, plasma cells, and lymphocytes infiltrating the alveolar septa. A perivascular and peribronchiolar lymphocytic infiltration was also present. Germinal center hypertrophy and multifocal lymphoid necrosis were observed in the lymph nodes. The heart and liver exhibited moderate fibrinosuppurative serositis. PCV3 mRNA was detected infrequently in macrophages in the alveolar wall (Fig. 1) and in histiocytes of the lymphoid follicles in the lymph nodes.
To investigate the genetic relationship of the PCV3 detected in this study with previously reported American, Brazilian, Italian, Chinese, and Korean isolates, sequencing of the full genome was conducted. The nucleotide and deduced amino acid (aa) sequences of the two PCV3 strains from the two farms were analyzed with BioEdit ver. 7.0.5.2 (Ibis Therapeutics, USA), and a phylogenetic tree was constructed using the maximum-likelihood method with MEGA 5.0 [17] and bootstrap analysis involving 1,000 replicates. The results indicated that all PCV3 strains used in this study were related to each other, and their sequences did not differ according to geographic location (Fig. 2). The nucleotide sequences (98.9–99.9%) and deduced aa sequences (98.6–100% for open reading frame 1 [ORF1] and 97.1–100% for ORF2) of the Korean PCV3 strains were most similar to those of viruses from the USA, Brazil, Italy, and China. The full genome of the two PCV3 strains, 16R927/2016 (GenBank No. MF063071) and P1705SCYC/2017 (GenBank No. MF063070), consisted of 2,000 nucleotides with 99.2% nucleotide sequence identity. ORF1 was 891 nucleotides long and encoded a 296 aa protein, and ORF2 was 645 nucleotides long and encoded a 214 aa protein.
This is the first report of clinical presentations of PCV3 infections in Korean pig herds. The pigs exhibited clinical symptoms, such as reproductive failure and respiratory distress, similar to those reported in the USA during the 2017 PCV3 infection period, with the exception of gross signs of PDNS.
We identified PCV3 DNA in the internal organs of aborted fetuses but did not detect any other virus or bacterium known to cause fetal damage despite the lack of microscopic evaluation of submitted autopsy samples, which may have hampered evaluation of the pathogenesis of PCV3. Moreover, we described the clinical presentation of abortion in Korean pig herds, which indicated that PCV3 might induce a reproductive disorder resembling that caused by PCV2. PCV2 has also been linked to reproductive disorders, late-term abortions, and farrowing, together with both stillborn and mummified piglets accompanied by myocarditis [210]. The second case of severe respiratory distress was due to a mixed infection with PRRSV, M. hyopneumoniae, S. suis, and PCV3, which resulted in bronchointerstitial pneumonia with fibrinosuppurative polyserositis. Furthermore, PCV3 was also detected by PCR and ISH analyses in the lung and lymph nodes. These results suggest that PCV3 was partially responsible for the pathological findings and respiratory signs; however, further study is needed to determine the role of PCV3 in pneumonia.
Multiple genotypes of PCV2 have circulated in Korean farms since PCV2 was identified in 1999 in pigs suffering from PMWS [3]. A genotype shift from PCV2a to PCV2b occurred around 2002, and PCV2d was first identified in Korea in a case of vaccine failure in 2013 [715]. Kwon et al. [8] reported that PCV2d was the predominant genotype in Korea nationwide and demonstrated that a genotype shift to PCV2d occurred before 2012. At present, multiple genotypes of PCV2 are co-circulating and PCV3 has been detected on Korean farms [9]. This could increase the likelihood of co-infection with different PCV genotypes in individual pigs, and/or alter the pathogenicity of PCV. Furthermore, the PCV2a-based vaccine available for use in Korean pigs is unlikely to be effective against PCV3 infection because the PCV3 capsid proteins have only 30% aa sequence identity with those of PCV2. Therefore, several factors may have led to the increased disease pattern complexity in Korean pigs. In conclusion, our results suggest that PCV3 might be a potential contributing cause of clinical disease in pigs, and further extensive studies are needed to cope effectively with PCV3-associated diseases in the future.
Figures and Tables
Fig. 1
Detection of porcine circovirus 3 mRNA (arrows) in lung alveolar macrophages by in situ hybridization. Scale bar = 100 µm.
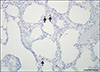
Fig. 2
Phylogenetic analysis of the complete genomes of two Korean porcine circovirus 3 (PCV3) strains and two reference strains, PCV2a-e and PCV3, available in GenBank. The PCV3 viruses used in this study are shown in red. The viruses in bold were detected in Korea. Numbers at branches represent bootstrap values greater than 50% of replicates.

Acknowledgments
This research was supported by a fund (project code No. B-1543069-2018-19-01) from the Animal and Plant Quarantine Agency, Korean Ministry of Agriculture, Food and Rural Affairs, and partly by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Golden Seed Project, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (213010-05-2-CG600 and PJ0128182018), Republic of Korea.
References
1. Barate AK, Lee HY, Jeong HW, Truong LQ, Joo HG, Hahn TW. An improved multiplex PCR for diagnosis and differentiation of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis. Korean J Vet Res. 2012; 52:39–43.

2. Brunborg IM, Jonassen CM, Moldal T, Bratberg B, Lium B, Koenen F, Schonheit J. Association of myocarditis with high viral load of porcine circovirus type 2 in several tissues in cases of fetal death and high mortality in piglets. A case study. J Vet Diagn Invest. 2007; 19:368–375.

3. Choi C, Chae C. In-situ hybridization for the detection of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Comp Pathol. 1999; 121:265–270.

4. Davies B, Wang X, Dvorak CM, Marthaler D, Murtaugh MP. Diagnostic phylogenetics reveals a new Porcine circovirus 2 cluster. Virus Res. 2016; 217:32–37.

5. Hamel AL, Lin LL, Nayar GP. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998; 72:5262–5267.

6. Kang I, Kim D, Han K, Seo HW, Oh Y, Park C, Lee J, Gottschalk M, Chae C. Optimized protocol for multiplex nested polymerase chain reaction to detect and differentiate Haemophilus parasuis, Streptococcus suis, and Mycoplasma hyorhinis in formalin-fixed, paraffin-embedded tissues from pigs with polyserositis. Can J Vet Res. 2012; 76:195–200.
7. Kim HH, Park SI, Hyun BH, Park SJ, Jeong YJ, Shin DJ, Chun YH, Hosmillo M, Lee BJ, Kang MI, Cho KO. Genetic diversity of porcine circovirus type 2 in Korean pigs with postweaning multisystemic wasting syndrome during 2005–2007. J Vet Med Sci. 2009; 71:349–353.

8. Kwon T, Lee DU, Yoo SJ, Je SH, Shin JY, Lyoo YS. Genotypic diversity of porcine circovirus type 2 (PCV2) and genotype shift to PCV2d in Korean pig population. Virus Res. 2017; 228:24–29.

9. Kwon T, Yoo SJ, Park CK, Lyoo YS. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet Microbiol. 2017; 207:178–180.

10. Madson DM, Patterson AR, Ramamoorthy S, Pal N, Meng XJ, Opriessnig T. Reproductive failure experimentally induced in sows via artificial insemination with semen spiked with porcine circovirus type 2. Vet Pathol. 2009; 46:707–716.

11. Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. 2017; 91:e01879–e01816.

12. Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, Deng X, Resende T, Vannucci F, Delwart E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016; 13:184.

13. Segales J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. 2012; 164:10–19.

14. Selim AM, Elhaig MM, Gaede W. Development of multiplex real-time PCR assay for the detection of Brucella spp., Leptospira spp. and Campylobacter foetus. Vet Ital. 2014; 50:269–275.
15. Seo HW, Park C, Kang I, Choi K, Jeong J, Park SJ, Chae C. Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea. Arch Virol. 2014; 159:3107–3111.

16. Stiles J, Prade R, Greene C. Detection of Toxoplasma gondii in feline and canine biological samples by use of the polymerase chain reaction. Am J Vet Res. 1996; 57:264–267.
17. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download