Abstract
Porcine deltacoronavirus (PDCoV) has emerged in several pig-raising countries and has been a causative pathogen associated with diarrheal diseases in South Korea since 2014. In the present study, we were able to isolate and cultivate a Korean PDCoV strain (KNU16-07) in cell culture and investigate its pathogenicity. PDCoV-inoculated piglets showed watery diarrhea accompanied by acute enteritis in the natural host. Sequencing analysis demonstrated the genetic stability of KNU16-07 for at least thirty serial passages.
Porcine deltacoronavirus (PDCoV) is a newly emerging enterotropic swine coronavirus that causes acute enteritis in nursing piglets [710]. PDCoV was first identified in Hong Kong, China in 2012 and, subsequently, has emerged in most major swine-producing nations in Asia and North America [111314]. The presence of PDCoV in South Korea was first announced in early 2014 [7]. Our previous report indicated that monoinfection by PDCoV and coinfection by PDCoV and porcine epidemic diarrhea virus (PEDV) are common in pig herds in South Korea [4].
In addition to studies on molecular surveillance and sequence analysis of PDCoV, more advanced PDCoV research is required for virus characterization in vitro and in vivo and to develop effective vaccines for PDCoV prevention. To accomplish these goals, obtaining a Korean PDCoV isolate that can grow efficiently in cell culture is essential. There have been no reports on the cultivation of a Korean PDCoV strain, although several research groups from the USA and China have reported the successful propagation of PDCoV strains in cell culture [123]. Therefore, the present study aimed to isolate PDCoV from PDCoV-positive fecal samples and assess the in vitro and in vivo PDCoV isolate KNU16-07.
Swine testicle (ST) cells (ATCC CRL-1746; ATCC, USA) were cultured in alpha minimum essential medium (α-MEM; Invitrogen, USA) containing 5% fetal bovine serum (Invitrogen) and antibiotic-antimycotic solutions (100×; Invitrogen). Small intestinal homogenates and stool specimens that tested PDCoV-positive by reverse transcription polymerase chain reaction (RT-PCR) [4] were selected for virus isolation experiments. PDCoV isolation was attempted on ST cells as described previously [3], but with the modification of adding trypsin (USB, USA) to the virus growth medium to obtain a final concentration of 5 to 10 µg/mL. The virus supernatants were purified and examined by using a transmission electron microscope (HT7700; Hitachi High Technologies, Japan) as described previously [9].
The PDCoV nucleocapsid (N) antigen was prepared from PK-PDCoV-N cells stably expressing the PDCoV N protein as described previously [8]. The cell lysate supernatant containing recombinant N protein was purified by using a HiTrap TALON crude column (GE Healthcare, USA) according to the manufacturer's instructions. The purified protein was concentrated with Amicon Ultra centrifugal filters 10K (Millipore, USA) and the final product was used as the antigen. PDCoV N-specific monoclonal antibodies (MAbs) were produced by immunizing 4-week-old male BALB/c mice as described previously [12].
The pig infection experiments described herein were performed at the KNU Animal Facility and followed guidelines established by its Institutional Animal Care and Use Committee (IACUC No. KNU-2016-2032) as described previously [611]. A total of four newborn piglets at 4 days of age were obtained from a commercial pig farm with no known prior PDCoV/PEDV outbreak or PEDV vaccination. All animals were determined to be free of PEDV as well as free of transmissible gastroenteritis virus and rotavirus. Pigs were randomly assigned to two experimental groups housed in two separated rooms: PDCoV-inoculated group (n = 3) in room 1 and sham-inoculated control group (n = 1) in room 2. Following a 1-day acclimation period, piglets in the challenge group were orally administered 1 mL of 105 TCID50/mL (TCID50, 50% tissue culture infective dose) of KNU16-07-P10 virus. The sham-inoculated pig was administered cell culture media as a placebo.
The complete genomic sequences of the KNU16-07 isolates at passage 5 (P5) and passage 10 (P10) were determined by performing rapid amplification of the cDNA ends and by using the traditional Sanger method as described previously [67]. The whole genomic nucleotide sequences of KNU16-07-P5 and KNU16-07-P10 were deposited in GenBank under accession numbers MG837130 and MG837131, respectively. The entire structural gene sequences of the isolate at passages 20 and 30 (KNU16-07-P20 and KNU16-07-P30) were also determined by the Sanger method, as described above, and deposited in the GenBank database under accession numbers MG837132 and MG837133, respectively.
The PDCoV isolate designated as KNU16-07 was isolated from the feces of a naturally infected piglet from a commercial farm located in Kyungpook Province. KNU16-07 virus produced distinct cytopathic effects (CPE) typical of a PDCoV infection, such as cell rounding, clumping together in clusters, and detachment in infected ST cells at passage 2. In later passages, visible CPE appeared at 12 h post-infection (hpi) and became pronounced by 24 hpi. Virus propagation was confirmed by an immunofluorescence assay using commercial (SD55-197; Medgene Labs, USA) and homemade (KDN4-1) PDCoV N-specific MAbs as described previously [6]. In contrast, no CPE or N-specific staining were evident in mock-inoculated cells (panel A in Fig. 1). Ultrastructural analysis of purified virus suspensions revealed multiple virus particles approximately 150 to 200 nm in diameter with typical spike-like surface projections that were morphologically indistinguishable from those of coronaviruses (panel B in Fig. 1). The viral genome levels in selected passages were examined by quantitative real-time RT-PCR as described previously [611]. The mean cycle threshold (Ct) value was determined to be 15.0, ranging from 13.9 (P10) to 16.1 (P5). The infectious titer of the isolate ranged from 107.8 to 108.8 TCID50/mL up to P5; this value was stably maintained in later passages. The peak viral titer reached 107.8 TCID50/mL or more beginning at P5 (panel C in Fig. 1). Analysis of growth kinetics demonstrated that KNU16-07 replicated rapidly and efficiently in ST cells, reaching a titer > 106 TCID50/mL by 12 hpi (panel D in Fig. 1).
The PDCoV-challenged piglets exhibited clinical signs including diarrheic feces and lethargy by 2 days post-inoculation (dpi) and then displayed vomiting and diarrhea for 3 days. All inoculated animals were positive for PDCoV, as determined by quantitative RT-PCR, at 1 or 2 dpi, and they shed PDCoV in feces. Peak mean Ct values of 16.8 (range, 16.7–16.8) and 18.7 (range, 18.5–18.9) were obtained at 3 to 4 dpi. The negative control pig remained active with normal feces and no fecal shedding of PDCoV detected throughout the study period. Two animals were independently euthanatized at 3 and 5 dpi for postmortem assessments, whereas the remaining piglets were necropsied at 35 dpi. Unlike the negative control piglet, all challenged piglets exhibited typical PDCoV-like macroscopic observations with thin and transparent small intestine walls (panel A in Fig. 2). The inoculated pigs also developed histopathological intestinal lesions associated with viral enteritis, including shortened and fused villi in the small intestine (panels B and C in Fig. 2). Furthermore, an immunohistochemistry assay detected PDCoV in the cytoplasm of villous enterocytes throughout the small intestines (panel D in Fig. 2). These macroscopic and microscopic observations are consistent with previous studies using USA PDCoV isolates [15]. Taken together, the results show that experimental infection of newborn piglets with PDCoV KNU16-07 induces clinical disease signs corresponding to acute enteritis, indicating enteropathogenicity of the isolate in the natural host. Additionally, all inoculated pigs had detectable PDCoV-specific neutralizing antibody (NA) by 14 dpi and maintained high NA titers of 1:128 to 1:256 thereafter, demonstrating the ability of KNU16-07 to induce efficient immune responses.
The entire genomes of the cell culture-adapted KNU16-07-P5 and -P10 strains were 25,422 nucleotides long and displayed genomic organization identical to all previously sequenced PDCoVs. The full-length genomic sequences of the original fecal sample (KNU16-07-feces, GenBank accession No. KY364365) and cell-passaged viruses were comparatively analyzed; the data are summarized in Table 1. Compared to KNU16-07-feces, KNU16-07-P5 and -P10 identically showed five nucleotide differences at positions 7737, 8218, 16481, 19808, and 21875. All mutations were non-synonymous, causing two, one, and two amino acid changes in the ORF1a, ORF1ab, and S coding regions, respectively. The two amino acid substitutions at positions 162 (Thr to Asn) and 851 (Asn to Ser) in the S coding region were sustained through passage 30 (KNU16-07-P30). No cell culture-adapting mutations arose in the E, M, and N protein-coding regions. These results reveal that the PDCoV isolate KNU16-07 is genetically stable during cell passages.
This is the first report describing the isolation, serial passages, in vitro and in vivo phenotypic features, and genotypic traits of a Korean PDCoV strain. The availability of this Korean isolate will enable cutting-edge research to investigate the molecular virology as well as to develop potential vaccines against emergent PDCoVs. The information presented in this study provides fundamental knowledge that should stimulate further basic and applied studies to improve our understanding of PDCoV.
Figures and Tables
Fig. 1
Cytopathology and growth properties of porcine deltacoronavirus (PDCoV) isolate KNU16-07 in infected swine testicle (ST) cells. (A) Cytopathic effect (CPE) formation in ST cells infected with PDCoV KNU16-07-P5 or -P10. PDCoV-specific CPEs were observed daily and were photographed at 24 h post-infection (hpi) using an inverted microscope. The infected cells were subjected to immunofluorescence assay using the indicated N-specific monoclonal antibodies and examined using a fluorescence microscope. (B) Electron microscopy images of PDCoV KNU16-07. Purified virions were negatively stained with 2% phosphotungstic acid and viewed by using transmission electron microscope. (C) Virus titers at selected passages. ST cells were infected with PDCoV KNU16-07 harvested at the indicated passage numbers. At 24 hpi, the virus supernatants were collected and virus titers were determined. (D) One-step growth kinetics for the KNU16-07 strain. At the indicated times post-infection, culture supernatants were harvested and virus titers determined. Results are expressed as mean values from triplicate wells; error bars represent SD. TCID50, 50% tissue culture infective dose; P, passage. 200× (A), 100,000× (B). Scale bar = 100 nm (B).
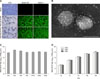
Fig. 2
Macroscopic and microscopic examinations of intestines of piglets inoculated with the Korean porcine deltacoronavirus (PDCoV) strain KNU16-07. (A) Small intestine of an inoculated pig at 3 days post-inoculation (dpi) showing thin and transparent intestinal walls (arrows) and an extended stomach filled with curdled milk. (B and C) H&E stained jejunum of a KNU16-07-inoculated pig at 3 dpi showing acute diffuse and severe atrophic enteritis with moderate vacuolation (arrows) of enterocytes lining the epithelium of atrophied villi. (D) Detection of PDCoV antigen by immunohistochemistry analysis of jejunum tissue sections from a virus-inoculated piglet at 3 dpi. The PDCoV antigen signals (arrows) appear red and were detected in villous epithelial cells. 100× (B and D), 200× (C).

Acknowledgments
This research was supported by a fund (project code No. Z-1543056-2016-17-01) of the Research of Animal and Plant Quarantine Agency, Republic of Korea.
References
1. Chen Q, Gauger P, Stafne M, Thomas J, Arruda P, Burrough E, Madson D, Brodie J, Magstadt D, Derscheid R, Welch M, Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015; 482:51–59.

2. Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol. 2016; 196:98–106.

3. Hu H, Jung K, Vlasova AN, Chepngeno J, Lu Z, Wang Q, Saif LJ. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J Clin Microbiol. 2015; 53:1537–1548.

4. Jang G, Lee KK, Kim SH, Lee C. Prevalence, complete genome sequencing and phylogenetic analysis of porcine deltacoronavirus in South Korea, 2014-2016. Transbound Emerg Dis. 2017; 64:1364–1370.

5. Jung K, Hu H, Eyerly B, Lu Z, Chepngeno J, Saif LJ. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg Infect Dis. 2015; 21:650–654.

6. Lee S, Kim Y, Lee C. Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU-141112. Virus Res. 2015; 208:215–224.

7. Lee S, Lee C. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2014; 2.

8. Lee S, Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015; 208:136–145.

9. Lee YN, Lee C. Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch Virol. 2013; 158:1765–1772.

10. Marthaler D, Jiang Y, Collins J, Rossow K. Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine deltacoronavirus from the United States. Genome Announc. 2014; 2:e00218-14.

11. Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K, Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis. 2014; 20:1347–1350.

12. Pan X, Kong N, Shan T, Zheng H, Tong W, Yang S, Li G, Zhou E, Tong G. Monoclonal antibody to N protein of porcine epidemic diarrhea virus. Monoclon Antib Immunodiagn Immunother. 2015; 34:51–54.

13. Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, Tantituvanont A, Kaewprommal P, Piriyapongsa J, Nilubol D. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis. 2017; 64:3–10.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download