Abstract
Recently, a novel atypical porcine pestivirus (APPV) in pig was reported. In this study, two APPV strains, APPV-China/GZ01/2016 (GZ01) and APPV-China/GD-SD/2016 (GD-SD), were identified in two newborn piglet herds with congenital tremor from China. The open reading frame of the two strains shared an 83.5% nucleotide identity. Phylogenetically, the APPV strains were placed into two groups: GZ01 belonged to group I and GD-SD belonged to group II. A high viral load was detected in the cerebellum (quantification cycles < 26). Further studies should be carried out to thoroughly elucidate the development of congenital tremors caused by APPV.
Congenital tremor (CT) in piglets is a well-known phenomenon characterized by tremors of the head and limbs. Based on the cause, CT is classified into one of two types of disease; type A or type B [4]. Type A can be further divided into five subgroups (types AI–AV) based on the cause of the disease [4]. While AIII and AIV are associated with piglets' genetic backgrounds, AI is caused by classical swine fever virus (CSFV) infection and AV is caused by metrifonate intoxication [4]. Type AII CT has long been regarded as a viral disease.
In 2015, a novel porcine pestivirus, atypical porcine pestivirus (APPV), was identified in the serum of pigs in the USA on the basis of metagenomic sequencing [6]. Subsequently, APPV was considered to be associated with the presence of CT in newborn piglets [1]. Widespread distribution of APPV was reported within type AII CT piglet herds in the USA, Germany, Netherlands, Spain, and Austria [231011]. Most recently, two reports described APPV circulating in Guangdong province, China [1415]. Moreover, CT disease has been duplicated by inoculating pregnant sows with APPV genome-positive serum [1].
The APPV is an enveloped virus belonging to the genus Pestivirus in the family Flaviviridae. The genome of APPV is a single-strand RNA of approximately 12 kb length that contains only one open reading frame (ORF) and encodes a polyprotein. Many pestiviruses cause severe clinical diseases, depending on the virus species and strain, such as CSFV and bovine viral diarrhea (BVD), which is caused by the highly virulent BVD virus II. In recent years, members of the Pestivirus genus have been reported in different animals including swine, rat, and the Rhinolophus affinis bat [5713].
Since July 2016, outbreaks of CT in newborn piglets have occurred in southern and southwestern China. Two reports have documented APPV-associated CT in southern China, but the prevalence of this novel virus in other areas of China was unclear [1415]. Since June 2016, a commercial pig farm containing 3,000 Large-White × Landrace sows in Guangdong province experienced an outbreak of CT in newborn piglets. Among the CT-affected piglets, morbidity was approximately 2.67% and mortality was 60%. The main clinical symptoms of the CT-affected piglets were head shaking and muscle tremor. Another commercial pig farm containing 800 Large-White × Landrace sows in Guizhou province experienced another CT outbreak; morbidity was approximately 50% during the epidemic period, but the mortality rate among the CT-affected piglets was unclear due to an absence of records. Three 3-day-old piglets with CT from each of these two pig farms were submitted to our laboratory for pathogen detection. Tissue suspensions prepared in phosphate-buffered saline were used for RNA extraction using TRIzol reagent (Invitrogen, USA) according to the manufacturer's instructions.
Presence of APPV was detected by performing one-step reverse transcription polymerase chain reaction (RT-PCR) using the PrimeScript One-Step RT-PCR Kit (Takara Bio, China) with a pair of specific primers (APPV-F: AGTC AACGGCAGGAGACCATC and APPV-R: GACCCTCACT GTTCCATCAAGCA). The RT-PCR conditions were: 50℃ for 30 min, 94℃ for 2 min, and then 30 cycles of 94℃ for 30 sec, 55℃ for 30 sec, and 72℃ for 50 sec, followed by final extension at 72℃ for 10 min. The PCR product size was approximately 660 bp. The obtained products were purified by using an Axygen AxyPrep DNA Gel Extraction Kit (Corning, USA) according to the manufacturer's instructions; after which, the products were cloned into the pMD-19T vector (Takara Bio) for sequencing. The purified recombinant plasmids were sequenced by Sangon Biotech (China). A BLASTN search in GenBank (National Center for Biotechnology Information, USA) was performed to compare the obtained nucleotide sequences with other sequences in the GenBank.
To sequence the two APPV strains, overlap RT-PCR was conducted via one-step RT-PCR with the PrimeScript One-Step RT-PCR Kit and primers designed at the conserved regions according to the APPV sequences in GenBank. However, 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE were not conducted to determine the ultimate 5′- and 3′-termini. The RT-PCR products were purified and cloned in the pMD19-T vector for sequencing, as described above. The sequences were assembled by using DNAStar software (DNAStar, USA), and the ORFs of the two strains were obtained and submitted to GenBank.
The ORF sequences of the viruses in this study, as well as other pestiviruses, were aligned by using the Clustal W method [9]. Phylogenic analysis was performed by using the maximum-likelihood method with tree topology and was verified by using 1,000 bootstrap replicates in MEGA software (ver. 7.0.26) [8].
To investigate tissue tropism of APPV-China/GZ2016 in CT piglets, a TaqMan quantitative real-time PCR targeting the NS5a encoding regions of APPV was developed based on GZ2016. The primers used were NS5a-F: CAAGCTGACCGATTATTG and NS5a-R: CCCATACAATGTCCCTAA. The probe used was the TaqMan NS5a-probe: FAM-TCATCCGTCAGGCTCT CAATAGTG-TAMRA. Total RNA from 11 varied tissue types from three APPV-China/GZ2016-affected piglets were extracted by using TRIzol reagent (Invitrogen) and were transcribed into cDNA by using a First-strand cDNA synthesis kit (Invitrogen). Real-time PCR was carried out by using TransStart Probe qPCR SuperMix (TransGen Biotech, China) on a Bio-Rad CFX Connect Real-Time PCR System. PCR reactions were performed on 2 µL of cDNA in a final volume of 20 µL that also contained 10 µL of 2× qPCR SuperMix, 0.4 µL each of the forward and reverse primers (final concentration 200 nM), and 0.4 µL of NS5a-probe (final concentration 200 nM). The PCR was performed as follows: 94℃ for 1 min, followed by 40 cycles of 94℃ for 10 sec and 60℃ for 30 sec.
An approximate 660 bp product was obtained by PCR from the CT-affected piglets, and the PCR results were negative for CSFV, porcine reproductive respiratory syndrome virus, and pseudorabies virus [12]. The products were sequenced and a BLASTN search was undertaken. The results showed that the nucleotide sequence obtained from the Guizhou piglets had 86% to 88% identity similarity to APPVs from the USA and Germany. However, the nucleotide sequence obtained from the Guangdong piglets had only 78% to 80% identity similarity to APPVs from the USA and Germany. The two sequences from the two different provinces in China shared only 77% identities; thus the APPV genomes from the two CT piglet herds from Guangdong and Guizhou province differed. The two strains were designated as APPV-China/GZ01/2016 (GZ01) and APPV-China/GD-SD/2016 (GD-SD).
The ORFs of GZ01 and GD-SD were obtained by performing overlap PCR. The obtained sequences are available under accession numbers KY475592 and KY475593. The ORFs of GZ01 and GD-SD comprised 10908 nucleotides (nt) encoding a 3592 amino acid (aa) polyprotein, and they shared 83.5% nucleotide identity. The ORF of GD-SD shared a 94.5% nucleotide identity (97.4%–97.5% aa identities) with two other APPV strains, GD1 (KX950761) and GD2 (KX950762), from Guangdong province [15]. The ORF of GZ2016 had nucleotide identities of 83.3% and 83.4% (92.1% and 92.2% aa identities) with GD1 and GD2, respectively. The ORF of GZ2016 had nucleotide identities of 87.9% to 90.8% (aa identities of 94.8%–95.7%) with APPVs from the USA, Germany, and Netherlands. However, the GD-SD strain only shared identities (83.1%–83.6%; aa identities of 91.4%–92.2%) with APPVs from the USA and Germany.
A phylogenetic tree was constructed by using MEGA software. The result showed that the APPV strains isolated in the study were divided into two groups. GZ01 clustered with the APPV from the USA and Germany (Fig. 1), which belonged to group I. However, GD-SD clustered with GD1, GD2, and GD3 from Guangdong of China, which belonged to group II.
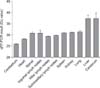
Tissue tropism was determined by performing real-time PCR. The results showed that high viral genome loads were found in the cerebellum (quantification cycles [Cq] < 26) and heart (Cq < 28.5) (Fig. 2). Lower viral genome loads were detected in the spinal cord, lung, spleen, kidney, submaxillary lymph nodes, inguinal lymph nodes, and hilar lymph nodes (Cq < 32), and the lowest viral genome load was detected in cerebrum and liver (Cq > 35) (Fig. 2).
In 2015, by using metagenomic sequencing, APPV was described as a novel pestivirus from the serum of pigs in the USA [6]. Subsequently, Arruda et al. [1] firstly detected APPV in samples of piglets with CT. Subsequently, the virus's genome was detected in Germany, Netherlands, Austria, and China [231011]. Widespread distribution of APPV in USA swine herds was determined by performing metagenomic sequencing and serum antibody detection [6]. But Arruda et al. [1] suggested that APPV was at a low prevalence in USA swine herds when its prevalence was assessed by using RT-PCR. A high prevalence of APPV was also reported in Germany [2]. The approximately 90% identity similarity of APPVs from these areas indicates that the genome of APPV is highly variable.
In this study, we identified two APPV from CT-affected piglets in southern and southwestern China by using RT-PCR; the results indicate that APPV is in circulation in China. The two strains shared only 83.5% nucleotide identity to each other. The GZ01 strain was clustered with the American and European APPV strains, whereas the GD-SD strain was genetically distant from those strains, indicating that the APPV has evolved for a long period of time epidemiologically [10]. Further investigation into the prevalence of APPV in China should be undertaken as soon as possible.
Although Arruda et al. [1] duplicated the clinical signs of CT by intrauterine infection of APPV, the transmission mechanism of APPV remains unclear. In this study, we investigated the tissue tropism of APPV. By using real-time PCR, a high viral genome load was detected in the cerebellum, which was consistent with results in other reports [110]. In contrast, Schwarz et al. [11] reported that high viral loads were detectable in the semen of APPV-infected young adult pigs, which suggested that sexual transmission might a significant APPV transmission route.
In summary, our results indicate that APPV has been circulating in pig breeding herds in China. This study provides an initial description of two APPV strains in China.
Figures and Tables
Fig. 1
Phylogenetic analysis of atypical porcine pestivirus (APPV) based on nucleotide sequencing of the viral open reading frame. The phylogenetic tree was constructed by performing maximum-likelihood analysis using MEGA software (ver. 7.0.26) [8] with 1,000 bootstrap replicates. The solid circle indicated the strains presented in this study. The APPV strains were placed into two groups: GZ01 belonged to group I and GD-SD belonged to group II.

Fig. 2
Tissue tropism of the atypical porcine pestivirus (APPV) strains in this study. Detection of APPV genome loads in different tissues of three three-day-old piglets with congenital tremor was accomplished by using quantitative reverse transcription polymerase chain reaction (qRT-PCR). Cq, quantification cycles.
Acknowledgments
This work was supported by the National Key Technologies R&D Program (2015BAD12B02-5), Guangdong Provincial Projects (2012A020200014 and 2013B020202002), and a Guangzhou City Project (201508020062).
References
1. Arruda BL, Arruda PH, Magstadt DR, Schwartz KJ, Dohlman T, Schleining JA, Patterson AR, Visek CA, Victoria JG. Identification of a divergent lineage porcine pestivirus in nursing piglets with congenital tremors and reproduction of disease following experimental inoculation. PLoS One. 2016; 11:e0150104.

2. Beer M, Wernike K, Dräger C, Höper D, Pohlmann A, Bergermann C, Schröder C, Klinkhammer S, Blome S, Hoffmann B. High prevalence of highly variable atypical porcine pestiviruses found in Germany. Transbound Emerg Dis. 2017; 64:e22–e26.

3. de Groof A, Deijs M, Guelen L, van Grinsven L, van Os-Galdos L, Vogels W, Derks C, Cruijsen T, Geurts V, Vrijenhoek M, Suijskens J, van Doorn P, van Leengoed L, Schrier C, van der Hoek. Atypical porcine pestivirus: a possible cause of congenital tremor type A-II in newborn piglets. Viruses. 2016; 8:pii: E271.

4. Done JT, Harding JD. Congenital tremor in pigs (trembling disease of piglets): lesions and causes. Dtsch Tierarztl Wochenschr. 1967; 74:333–336. German.
5. Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, Conte JM, Ng J, Garcia J, Bhuva NP, Lee B, Che X, Quan PL, Lipkin WI. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio. 2014; 5:e01933–e01914.

6. Hause BM, Collin EA, Peddireddi L, Yuan F, Chen Z, Hesse RA, Gauger PC, Clement T, Fang Y, Anderson G. Discovery of a novel putative atypical porcine pestivirus in pigs in the USA. J Gen Virol. 2015; 96:2994–2998.

7. Kirkland PD, Frost MJ, Finlaison DS, King KR, Ridpath JF, Gu X. Identification of a novel virus in pigs—Bungowannah virus: a possible new species of pestivirus. Virus Res. 2007; 129:26–34.

8. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1874.

9. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23:2947–2948.

10. Postel A, Hansmann F, Baechlein C, Fischer N, Alawi M, Grundhoff A, Derking S, Tenhündfeld J, Pfankuche VM, Herder V, Baumgärtner W, Wendt M, Becher P. Presence of atypical porcine pestivirus (APPV) genomes in newborn piglets correlates with congenital tremor. Sci Rep. 2016; 6:27735.

11. Schwarz L, Riedel C, Högler S, Sinn LJ, Voglmayr T, Wöchtl B, Dinhopl N, Rebel-Bauder B, Weissenböck H, Ladinig A, Rümenapf T, Lamp B. Congenital infection with atypical porcine pestivirus (APPV) is associated with disease and viral persistence. Vet Res. 2017; 48:1.

12. Wang PP, Dong JG, Zhang LY, Liang PS, Liu YL, Wang L, Fan FH, Song CX. Sequence and phylogenetic analyses of the Nsp2 and ORF5 genes of porcine reproductive and respiratory syndrome virus in boars from South China in 2015. Transbound Emerg Dis. 2017; 64:1953–1964.

13. Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, Zhang J, Dong J, Sun L, Du J, Liu L, Xue Y, Wang J, Yang F, Zhang S, Jin Q. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J Virol. 2012; 86:10999–11012.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download