Abstract
Figures and Tables
Fig. 1
Histopathological lesions of the pancreas from mice inoculated with CVB2 on days 7 (A, C, E, and G) and 13 (B, D, F, and H) post-inoculation (pi). Mice were mock-infected with saline (A and B), or inoculated with CB2/04/243 (C and D), CB2/04/279 (E and F), or CVB/O (G and H). Mice were sacrificed on days 7 and 13 pi and pancreatic tissues were fixed in formalin, paraffin embedded, cross-sectioned at a 3 µm thickness, and stained with hematoxylin and eosin. 200×.
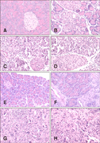
Fig. 2
Mice inoculated with CB2/04/243( ) were compared with those inoculated with CVB/O (■) and CB2/04/279 (□) for pancreatitis severity level on days 7 and 13 post-inoculation (pi). Average severity grades of lesions in animals: 1, minimal; 2, mild; 3, moderate; 4, marked. Severity grades among strains were compared by independent t-tests (n = 6 or 8 per group). *p < 0.05 considered statistically significant.
) were compared with those inoculated with CVB/O (■) and CB2/04/279 (□) for pancreatitis severity level on days 7 and 13 post-inoculation (pi). Average severity grades of lesions in animals: 1, minimal; 2, mild; 3, moderate; 4, marked. Severity grades among strains were compared by independent t-tests (n = 6 or 8 per group). *p < 0.05 considered statistically significant.

Fig. 3
Histopathological lesions of the hearts from mice inoculated with CVB2 on days 7 (A, C, E, and G) and 13 (B, D, F, and H) post-inoculation (pi). Mice were mock-infected with saline (A and B), or inoculated with CB2/04/243 (C and D), CB2/04/279 (E and F), or CVB/O (G and H). Mice were euthanized 7 and 13 days pi. Arrows indicate area of inflammation in heart. Hearts were fixed in formalin, paraffin embedded, cross-sectioned at a 3 µm thickness, and stained with hematoxylin and eosin. 100×.
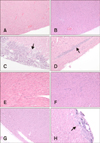
Table 1
Summary of virology and pathology results

To compare changes of identity, divergence of full genome sequence, and histology between CVB2 strains, the following was undertaken. Mice were inoculated intraperitoneally with 2.5 × 104 PFU/0.5 mL (PFU, plaque-forming unit) of a CVB2 strain to reveal histopathological changes in their heart and pancreas on days 7and 13 post-inoculation (pi). In addition, percentages of identity and divergence of homology of genome sequences between the CB2 reference strain (CVB/O) and the Korean isolates, which differed in clinical signs, were compared. The sequences of Korean isolate strains CB2/04/279 and CB2/04/243 had a high degree of homology in contrast, strains CB2/04/279 and CVB/O exhibited highly similar patterns of histological change in mice. Positive (+) and negative (−) of the histological lesion in mice group. *Percent nucleotide sequence identity between full genomes of two CB2 strains. †Percent nucleotide sequence divergence between full genomes of two CB2 strains.
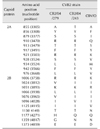
Table 2
Differences of amino acid of CVB2 Korean strains (CB2/04/279, CB2/04/243) and reference strain (CVB/O) in P1 (VP1–VP4)
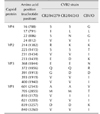
Table 3
Differences of amino acid of CVB2 Korean strains (CB2/04/279, CB2/04/243) and reference strain (CVB/O) in P2 (2A–2C)




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download