Abstract
The present study investigated the prevalence and mechanisms of fluoroquinolone (FQ)/quinolone (Q) resistance in Escherichia (E.) coli isolates from companion animals, pet-owners, and non-pet-owners. A total of 63 E. coli isolates were collected from 104 anal swab samples, and 27 nalidixic acid (NA)-resistant isolates were identified. Of those, 10 showed ciprofloxacin (CIP) resistance. A plasmid-mediated Q resistance gene was detected in one isolate. Increased efflux pump activity, as measured by organic solvent tolerance assay, was detected in 18 NA-resistant isolates (66.7%), but was not correlated with an increase in minimum inhibitory concentration (MIC). Target gene mutations in Q resistance-determining regions (QRDRs) were the main cause of (FQ)Q resistance in E. coli. Point mutations in QRDRs were detected in all NA-resistant isolates, and the number of mutations was strongly correlated with increased MIC (R = 0.878 for NA and 0.954 for CIP). All CIP-resistant isolates (n = 10) had double mutations in the gyrA gene, with additional mutations in parC and parE. Interestingly, (FQ)Q resistance mechanisms in isolates from companion animals were the same as those in humans. Therefore, prudent use of (FQ)Q in veterinary medicine is warranted to prevent the dissemination of (FQ)Q-resistant bacteria from animals to humans.
Quinolone (Q) and fluoroquinolone (FQ) are broad-spectrum synthetic antimicrobials used to treat bacterial infections in humans and animals [2327]. Since they are very potent antimicrobial agents against Gram-negative bacteria, including Escherichia (E.) coli, these agents have been widely used to treat a range of infections in human and veterinary medicine. Consequently, (FQ)Q resistance has markedly increased worldwide, posing a significant threat to the health of animals and humans [2327].
Three major mechanisms of (FQ)Q resistance have been reported: (1) mutations in genes encoding DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) that are associated with quinolone resistance-determining regions (QRDRs); (2) the presence of plasmid-mediated Q resistance (PMQR) genes; and (3) reduced accumulation of drugs or chemicals due to active efflux pump activity [17]. PMQR genes include members of the qnr gene family (qnrA, qnrB, and qnrS) as well as genes encoding FQ-modifying enzyme [aac-(6′)-Ib-cr] and the efflux pump (qepA) [40]. AcrAB-TolC overexpression is a major resistance mechanism against (FQ)Q that is associated with increased efflux pump activity and contributes to multi-drug resistance (MDR) in E. coli [28]. AcrAB-TolC has three components: a transporter of the resistance-nodulation-division family (AcrB), a periplasmic accessory protein (AcrA), and an outer membrane protein (TolC) [28].
E. coli are usually commensal bacteria in humans and animals. They are also considered major causative agents of bacterial infections. E. coli easily acquire antimicrobial resistance by genetic mutation and horizontal gene transfer [3]. The transmission of antimicrobial-resistant E. coli from animals to humans has been demonstrated [30]. Several studies have investigated multi-factorial (FQ)Q resistance mechanisms in E. coli isolated from humans and food-producing animals [19]. However, few studies have examined the prevalence and the resistance mechanisms of (FQ)Q-resistant E. coli from companion animals [13], and most of these have been limited to elucidating one or two of the above-mentioned (FQ)Q resistance mechanisms. Moreover, there have been no studies investigating the distribution of (FQ)Q-resistant E. coli in pets and pet-owners. Clarifying how (FQ)Q resistance develops and distributes in companion animals and the humans that they contact is important for understanding (FQ)Q resistance trends in veterinary medicine. To this end, the present study examined the frequency of nalidixic acid (NA)-resistant E. coli isolated from companion animals and their owners and investigated the three basic mechanisms of (FQ)Q resistance in these isolates relative to those obtained from non-pet-owners.
Sampling was carried out with informed consent from owners of companion animals and other human subjects. A total of 104 anal swab samples were collected from four local veterinary clinics, one veterinary teaching hospital, and one local university in Seoul, Korea between April 2010 and November 2012. The sampling procedures that were used have been previously described [14]. Swab samples were obtained from 49 dogs, four cats, 14 dog owners, three cat owners and 34 non-pet-owners (Table 1). People living with and without pets at the time of sampling were designated as pet-owners and non-pet-owners, respectively. Owners were selected from among visitors of four local veterinary clinics and a veterinary teaching hospital; non-pet-owners were selected from among freshman students at a university. All protocols and procedures were approved by the institutional review board at the Seoul National University (IRB No. 1208/001-004).
E. coli isolation and confirmation was carried out as previously described [6]. After isolating and identifying E. coli from swab samples, 30-µg NA antimicrobial disks (BD Biosciences, USA) were used to select NA-resistant isolates according to Clinical and Laboratory Standards Institute (CLSI) standards [7].
The susceptibility of 27 NA-resistant isolates to other antimicrobials was characterized by using the following antimicrobial disks (BD Biosciences, USA): ampicillin (AM; 10 µg), amoxicillin/clavulanic acid (AMC; 20/10 µg), ceftazidime (CAZ; 30 µg), cefotetan (CTT; 30 µg), imipenem (10 µg), gentamicin (10 µg), tetracycline (30 µg), ciprofloxacin (CIP; 5 µg), sulfamethoxazole/trimethoprim (1.25/23.75 µg), chloramphenicol (C; 30 µg), aztreonam (ATM; 30 µg), ceftriaxone (CRO; 30 µg), and cefotaxime (CTX; 30 µg). Susceptibility or resistance to antimicrobials was determined according to CLSI standards [7]. E. coli ATCC 25922 was used as a reference strain (American Type Culture Collection, USA). MDR isolates were defined as isolates showing resistance to more than three different classes of antimicrobials [21].
NA-resistant E. coli isolates showing inhibition zone diameters of ≤ 25 mm against CRO were selected for use in the confirmation test for extended spectrum β-lactamase (ESBL) production [7]. Isolates were identified as ESBL-producing E. coli by applying the disk diffusion method using CAZ, CAZ/clavulanic acid (CL) (30/10 µg), CTX, and CTX/CL (30/10 µg); those isolates showing resistance to CAZ and/or CTX in combination with an increase in inhibition zone diameter of ≥ 5 mm for CAZ/CL and/or CTX/CL were defined as ESBL-producing E. coli [33]. In addition, the presence of the blaCTX-M gene in ESBL-producing isolates was determined by performing polymerase chain reaction (PCR) with CTX-M universal primers [22].
The MICs of NA and CIP were determined for the 27 NA-resistant E. coli isolates by using the broth microdilution method according to CLSI standards [7], with E. coli ATCC 25922 used as a reference strain.
The 27 NA-resistant E. coli isolates were screened by PCR for the presence of the following PMQR genes: aac(6′)-Ib-cr, qepA, qnrA, qnrB, and qnrS [53639]. The cr variant of the aac(6′)-Ib gene was identified by direct sequencing of the amplified aac(6′)-Ib gene [26]. Mutations in DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) genes of the NA-resistant isolates were identified by using specific primers [2103237]. The wild-type E. coli K-12 sequence (GenBank accession No. U00096) was used as a reference [19].
The activity of the AcrAB-TolC efflux pump system in NA-resistant E. coli isolates was measured by using the OST assay [35]. For efficiency-of-plating assays [35], cultures of isolates in logarithmic growth phase were diluted to an optical density of 0.2 at a wavelength of 530 nm, and 100-µL aliquots were spread onto Luria-Bertani (LB) agar, which was then overlaid with a mixture of hexane and cyclohexane [3:1 (v/v)]. The plates were sealed and incubated for 24 to 36 h at 30℃. The number of colonies was counted in triplicate and colony growth was recorded as confluent (++, ≥ 100 colonies), visible (+, < 100 colonies), or none (−). E. coli ATCC 25922 was used as a reference strain.
To assess the relevance of the three (FQ)Q resistance mechanisms to the increase in (FQ)Q resistance in E. coli, we evaluated the frequencies of point mutations in the QRDR region and PMQR genes as well as efflux pump activity in the 27 NA-resistant E. coli isolates. To assess the effects of point mutations and efflux pump activity, an additional comparison was made by determining Pearson's correlation coefficient (R) [1]. The strength of efflux pump activity was graded based on the result of OST test described above (−, 0; +, 1; ++, 2). The R values were calculated between the number of point mutations or strength of efflux pump activity and MICs of NA or CIP by using SPSS software (ver. 23; IBM, USA) [1].
A total of 63 E. coli isolates (60.6%) were collected from 104 anal swab samples. Of these, 27 isolates (42.9%) were determined to be NA-resistant E. coli. Ten isolates (15.9%) were resistant to CIP, all of which also showed resistance to NA (Table 1). Overall, E. coli isolation and (FQ)Q resistance rates were higher in non-pet-owners than in pet-owners (Table 1).
Of the 27 NA-resistant E. coli isolates, 23 (85.2%) were resistant to at least one additional antimicrobial (data not shown) and 20 (74.1%) were identified as MDR (Table 2). Antibiogram analysis revealed that more than half of NA-resistant isolates were also resistant to AM (17/27, 63.0%), AMC (17/27, 63.0%), and TE (14/27, 51.9%). Ten of the NA-resistant isolates showed resistance to CIP (10/27, 37.0%); interestingly, these were all identified as MDR (Table 2). On the contrary, resistances against CAZ, CTT, C, ATM, CRO, and CTX were relatively low. In addition, 3 of the 27 NA-resistant isolates (11.1%) were identified as ESBL-producing E. coli harboring the blaCTX-M gene and were obtained from two dogs and a non-pet-owner (data not shown).
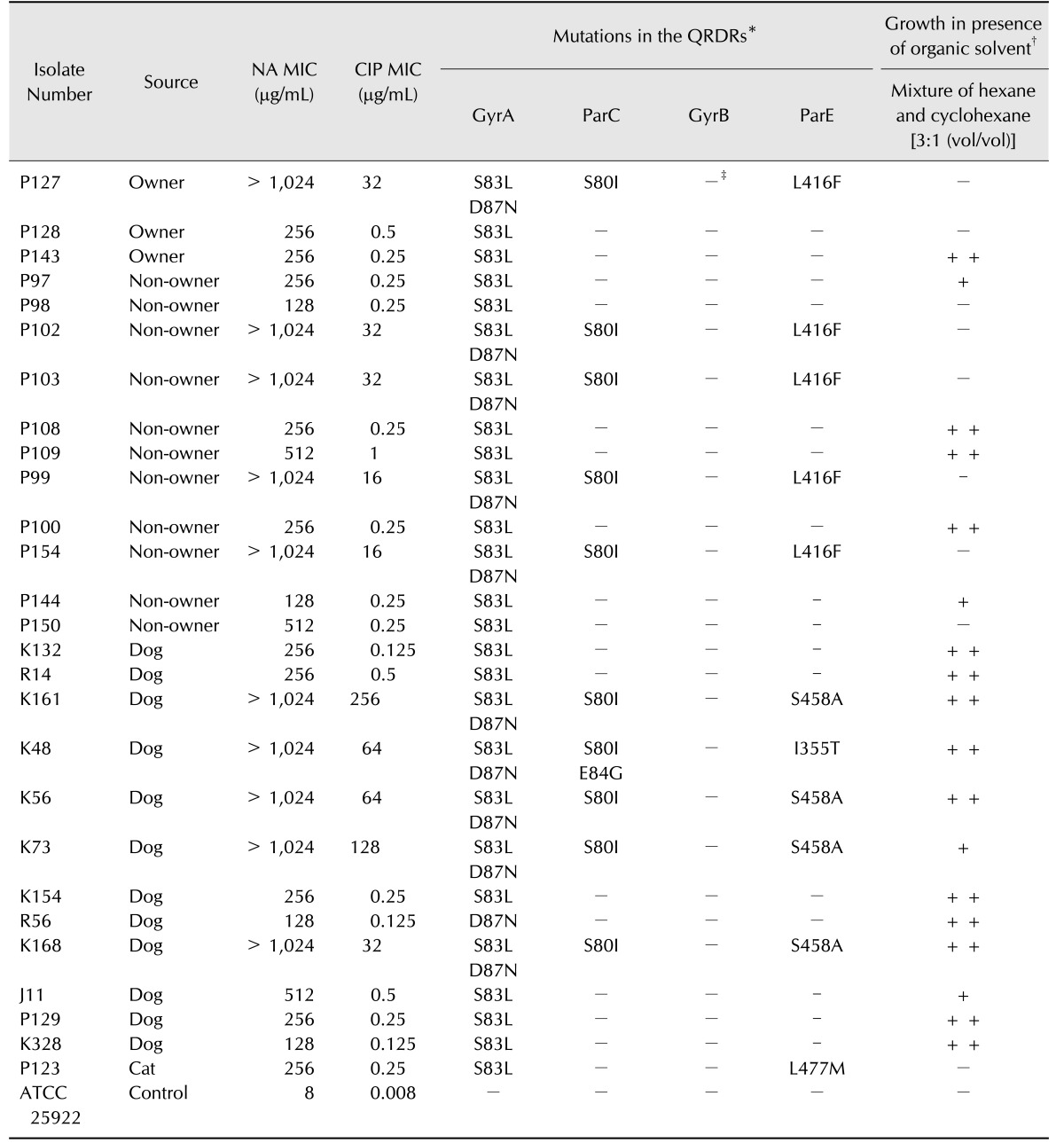
The MICs of NA for the 27 NA-resistant E. coli isolates ranged from 128 to > 1024 µg/mL. Ten isolates showed much higher MICs of NA (> 1024 µg/mL) than the others, and only those isolates showed resistance to CIP (MICs of CIP, 16–256 µg/mL). The remaining isolates showed low MICs of CIP, ranging from 0.125 to 1 µg/mL (Table 3).
Mutations were identified as nucleotide alterations in QRDRs responsible for changes in amino acid sequences in the protein products. Mutations were detected in the gyrA gene in all NA-resistant isolates (Table 3); 10/27 (37.0%) had double amino acid substitutions (S83L and D87N); 16/27 (59.3%) had a single amino acid substitution (S83L); and 1/27 (3.7%) had a single D87N substitution. Mutations in the parC gene were detected in 10/27 isolates (37.0%); nine (33.3%) had a single S80I mutation, and one had double mutations of S80I and E84G. However, no mutations were found in the gyrB gene. Eleven isolates (40.7%) had mutations in codons 355, 416, 458, or 477 of the parE gene (Table 3).
Notably, all 10 isolates with double amino acid substitutions in GyrA had point mutations in both parC and parE genes, and only these isolates were resistant to CIP. While parC mutations were detected only in these 10 isolates, parE mutations were found in those 10 isolates and an additional isolate with a single amino acid substitution in the gyrA gene (No. P123; Table 3). Of the 10 CIP-resistant isolates, five originating from humans had an amino acid substitution in only codon 416 of parE (L416F). In contrast, the other five CIP-resistant E. coli isolates from dogs had amino acid substitutions in codon 355 or 458 of parE (I355T or S458A) (Table 3). A PMQR gene was detected in only one isolate (No. K73), which harbored the aac(6′)-Ib-cr gene encoding a CIP-modifying enzyme (data not shown).
Efflux pump activity in the 27 NA-resistant E. coli isolates was measured by using the OST assay as described in the Materials and Methods section. A total of 14 (51.9%) and 4 (14.8%) isolates showed confluent and visible growth, respectively, on the organic solvent mixture (Table 3).
We analyzed the correlations between efflux pump activity or target mutations and MICs of NA or CIP in the 27 NA-resistant isolates and observed that the MICs of both NA and CIP were highly correlated with the number of point mutations in the QRDR (R = 0.878 and 0.954, respectively; Fig. 1). However, efflux pump activity was not correlated with either the NA MIC (R = −0.239) or the CIP MIC (R = −0.169) in NA-resistant E. coli isolates (Fig. 1).
Many epidemiological studies have reported that companion animals in households are potential sources of transmissible bacteria such as E. coli and Salmonella spp. (reviewed in [9]). However, in the present study, the isolation and antimicrobial resistance rates of E. coli were not higher in pet-owners than in non-pet-owners. Another study reported a lower risk of MDR staphylococci carriage in nursing home residents living with companion animals than in those living without pets [12]. These findings may suggest that although companion animals can be sources of bacterial infections in households, they do not always negatively affect the hygienic status of their owners.
(FQ)Q resistance is closely associated with MDR in E. coli [31]. We observed that about 74% of NA-resistant E. coli isolates were MDR. Cross-antimicrobial resistance between Q and β-lactams frequently occurs in E. coli and Klebsiella spp. due to the extensive use of antimicrobials against these bacteria in human and veterinary medicine [11]. The mechanism underlying the association between MDR and Q resistance is currently unclear. In our study, the MDR rate was higher in CIP-resistant than in NA-resistant E. coli isolates (100% vs. 74.1%). As shown in previous studies [131519], as well as in the current study, CIP resistance is strongly correlated with multiple mutations in QRDRs.
In this study, all 27 NA-resistant E. coli had at least one target mutation in the QRDRs. Low or high efflux pump activity was observed in 18 of those isolates (66.7%), but only one (3.7%) harbored a PMQR gene. Mobile (FQ)Q resistance gene transfer by plasmids has been demonstrated, and high detection rates for PMQR genes in (FQ)Q-resistant E. coli isolated from human and animal specimens have been reported [2324]. However, other studies have reported low detection rates or an absence of PMQR genes in FQ-resistant E. coli from animals [1319]. The results of the present study are consistent with the latter and highlight the low prevalence of PMQR in Korea. The presence of PMQR genes is closely associated with that of genes encoding ESBL in E. coli isolates [18]. Accordingly, the isolate harboring aac-(6′)-Ib-cr (No. K73) was determined to be an ESBL-producing E. coli strain carrying the blaCTX-M gene in this study.
We found no correlation between increased efflux pump activity and increases in MICs of both NA and CIP in NA-resistant E. coli. When MICs in 15 isolates harboring the same single amino acid substitution in GyrA (S83L) were compared, to exclude other mutational variables in QRDRs, MICs in isolates with high OST were not higher than those in isolates with no OST (average MICs: NA, 277 vs. 299 µg/mL and CIP, 0.33 vs. 0.33 µg/mL). Efflux pumps reduce the concentration of substrates within cells via active transport, and an increase in their activity has been reported to contribute to reduced (FQ)Q susceptibility [27]. However, our results indicate that efflux pump activity did not contribute to resistance against (FQ)Q in these resistant isolates. If mutations in QRDRs lead to sufficiently high levels of (FQ)Q resistance in E. coli, the activity of the efflux pump may not further increase the MICs of (FQ)Q [1]. Similar findings were reported in other Gram-negative bacteria such as Klebsiella pneumoniae and Campylobacter spp. [829]. In contrast, the number of mutations in QRDRs was strongly correlated with increases in the MICs of both NA and CIP. Particularly, in QRDRs the (FQ)Q resistance in clinical E. coli isolates is more closely associated with mutations in the gyrA gene, whereas mutations in gyrB, parC, and parE genes are less important in the establishment of (FQ)Q resistance [16]. A single mutation in the gyrA gene has been linked to low FQ resistance in E. coli [38], while high FQ resistance was reported to be acquired via accumulation of mutations in QRDRs [15]. In particular, FQ resistance was related to double amino acid substitutions in GyrA with or without mutations in parC and parE genes [131920]. In the present study, all CIP-resistant isolates from both humans and animals had double point mutations in gyrA concurrent with target mutations in parC and parE genes. These results indicate that (FQ)Q resistance mechanisms in E. coli at animal hospitals are similar to those observed in humans.
Mutations in the parC or parE gene have been reported to be closely related to secondary mutations in the gyrA gene [420]. Consistent with those results, all mutations in parC or parE were detected in isolates with double amino acid substitutions, except for one case. Among the four amino acid substitutions in ParE identified in this study, three have been previously reported (I355T, L416F, and S458A) [1925]. However, the amino acid substitution (L477M) found in a cat isolate (No. P123) is a novel finding. Interestingly, the patterns of parE mutations observed in the NA-resistant E. coli isolates were distinct in each species; L477M and L416F were present only in cat and human isolates, respectively. All I355T and S458A substitutions were only found in dog isolates, and they were in conjunction with amino acid substitutions in both GyrA and ParC. Taken together, the CIP-resistant isolates from humans were distinct from those obtained from dogs and cats in terms of the position of the parE gene mutations.
In conclusion, this is the first study to examine the mechanisms of (FQ)Q resistance in NA-resistant E. coli isolates from companion animals and their owners compared to those from non-pet-owners. The rates of E. coli isolation and (FQ)Q resistance were not higher in pet-owners than in non-pet-owners, which may suggest that persons living with pets are not always at a higher risk of bacterial infection than those living without pets. The prevalence of PMQR genes was very low and efflux pump activity was not found to contribute alone to the acquisition of high-level (FQ)Q resistance. Target site alterations in QRDRs appeared to be the most important mechanism contributing to high-level (FQ)Q resistance in E. coli of both animal and human origins in Korea. Since the (FQ)Q resistance mechanisms in pet isolates were the same as those found in human isolates, prudent use of (FQ)Q by veterinarians is warranted in order to prevent the development and dissemination of (FQ)Q-resistant bacteria. To this end, a continuous monitoring process in veterinary hospitals may be needed to identify the trends and dissemination of (FQ)Q resistance in companion animals and their owners.
Acknowledgments
The study was partly supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the High Value-added Food Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (No. 1150043). Additional support was provided by the project titled “Evaluation of food safety and development of novel processed meat products for application of deep sea water”, funded by the Ministry of Oceans and Fisheries, Republic of Korea.
References
1. Aathithan S, French GL. Organic solvent tolerance and fluoroquinolone resistance in Klebsiella pneumoniae clinical isolates. J Antimicrob Chemother. 2009; 64:870–871. PMID: 19679596.
2. Bai H, Du Jf, Hu M, Qi J, Cai YN, Niu WW, Liu YQ. Analysis of mechanisms of resistance and tolerance of Escherichia coli to enrofloxacin. Ann Microbiol. 2012; 62:293–298.
3. Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol. 2008; 153(Suppl 1):S347–S357. PMID: 18193080.

4. Breines DM, Ouabdesselam S, NG EY, Tankovic J, Shah S, Soussy CJ, Hooper DC. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob Agents Chemother. 1997; 41:175–179. PMID: 8980775.
5. Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007; 60:394–397. PMID: 17561500.

6. Chung YS, Song JW, Kim DH, Shin S, Park YK, Yang SJ, Lim SK, Park KT, Park YH. Isolation and characterization of antimicrobial-resistant E. coli from national horse racetracks and private horse-riding courses in Korea. J Vet Sci. 2016; 17:199–206. PMID: 26645344.
7. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Nineteenth Informational Supplement. CLSI document M100-S19. Wayne: Clinical and Laboratory Standards Institute;2009.
8. Corcoran D, Quinn T, Cotter L, Fanning S. Relative contribution of target gene mutation and efflux to varying quinolone resistance in Irish Campylobacter isolates. FEMS Microbiol Lett. 2005; 253:39–46. PMID: 16213669.
9. Doyle MP, Ruoff KL, Pierson M, Weinberg W, Soule B, Michaels BS. Reducing transmission of infectious agents in the home-part I: sources of infection. Dairy Food Environ Sanit. 2000; 20:330–337.
10. Fendukly F, Karlsson I, Hanson HS, Kronvall G, Dornbusch K. Patterns of mutations in target genes in septicemia isolates of Escherichia coli and Klebsiella pneumoniae with resistance or reduced susceptibility to ciprofloxacin. APMIS. 2003; 111:857–866. PMID: 14510643.
11. Frank T, Mbecko JR, Misatou P, Monchy D. Emergence of quinolone resistance among extended-spectrum beta-lactamase-producing Enterobacteriaceae in the Central African Republic: genetic characterization. BMC Res Notes. 2011; 4:309. PMID: 21867486.

12. Gandolfi-Decristophoris P, De Benedetti A, Petignat C, Attinger M, Guillaume J, Fiebig L, Hattendorf J, Cernela N, Regula G, Petrini O, Zinsstag J, Schelling E. Evaluation of pet contact as a risk factor for carriage of multidrug-resistant staphylococci in nursing home residents. Am J Infect Control. 2012; 40:128–133. PMID: 21824684.
13. Guillard T, de Jong A, Limelette A, Lebreil AL, Madoux J, de Champs C. The ComPath Study Group. Characterization of quinolone resistance mechanisms in Enterobacteriaceae recovered from diseased companion animals in Europe. Vet Microbiol. 2016; 194:23–29. PMID: 26701806.
14. Gustavsson L, Westin J, Andersson LM, Lindh M. Rectal swabs can be used for diagnosis of viral gastroenteritis with a multiple real-time PCR assay. J Clin Virol. 2011; 51:279–282. PMID: 21683649.

15. Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994; 38:1284–1291. PMID: 8092826.
16. Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005; 25:358–373. PMID: 15848289.
17. Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005; 41(Suppl 2):S120–S126. PMID: 15942878.

18. Jiang Y, Zhou Z, Qian Y, Wei Z, Yu Y, Hu S, Li L. Plasmid-mediated quinolone resistance determinants qnr and aac(6′)-Ib-cr in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in China. J Antimicrob Chemother. 2008; 61:1003–1006. PMID: 18299311.
19. Karczmarczyk M, Martins M, Quinn T, Leonard N, Fanning S. Mechanisms of fluoroquinolone resistance in Escherichia coli isolates from food-producing animals. Appl Environ Microbiol. 2011; 77:7113–7120. PMID: 21856834.
20. Khodursky AB, Zechiedrich EL, Cozzarelli NR. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci U S A. 1995; 92:11801–11805. PMID: 8524852.
21. Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, Godfrey P, Palmer KL, Bodi K, Mongodin EF. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012; 3:e00112-12. PMID: 22617140.

22. Liebana E, Batchelor M, Hopkins K, Clifton-Hadley F, Teale C, Foster A, Barker L, Threlfall E, Davies R. Longitudinal farm study of extended-spectrum β-lactamase-mediated resistance. J Clin Microbiol. 2006; 44:1630–1634. PMID: 16672386.

23. Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, Lü D, Huang L, Zhang Y, Liu J. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother. 2009; 53:519–524. PMID: 18936192.
24. Morgan-Linnell SK, Boyd LB, Steffen D, Zechiedrich L. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009; 53:235–241. PMID: 18838592.
25. Nam YS, Cho SY, Yang HY, Park KS, Jang JH, Kim YT, Jeong JW, Suh JT, Lee HJ. Investigation of mutation distribution in DNA gyrase and topoisomerase IV genes in ciprofloxacin-non-susceptible Enterobacteriaceae isolated from blood cultures in a tertiary care university hospital in South Korea, 2005–2010. Int J Antimicrob Agents. 2013; 41:126–129. PMID: 23265914.
26. Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother. 2006; 50:3953–3955. PMID: 16954321.
27. Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014; 22:438–445. PMID: 24842194.

28. Sato T, Yokota Si, Okubo T, Ishihara K, Ueno H, Muramatsu Y, Fujii N, Tamura Y. Contribution of the AcrAB-TolC efflux pump to high-level fluoroquinolone resistance in Escherichia coli isolated from dogs and humans. J Vet Med Sci. 2013; 75:407–414. PMID: 23149545.
29. Schneiders T, Amyes S, Levy S. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob Agents Chemother. 2003; 47:2831–2837. PMID: 12936981.
30. So JH, Kim J, Bae IK, Jeong SH, Kim SH, Lim SK, Park YH, Lee K. Dissemination of multidrug-resistant Escherichia coli in Korean veterinary hospitals. Diagn Microbiol Infect Dis. 2012; 73:195–199. PMID: 22516765.
31. Strand L, Jenkins A, Henriksen IH, Allum AG, Grude N, Kristiansen BE. High levels of multiresistance in quinolone resistant urinary tract isolates of Escherichia coli from Norway; a non clonal phenomen? BMC Res Notes. 2014; 7:376. PMID: 24941949.

32. Vila J, Ruiz J, Goñi P, De Anta M. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996; 40:491–493. PMID: 8834907.
33. von Salviati C, Laube H, Guerra B, Roesler U, Friese A. Emission of ESBL/AmpC-producing Escherichia coli from pig fattening farms to surrounding areas. Vet Microbiol. 2015; 175:77–84. PMID: 25465658.
34. Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001; 45:1515–1521. PMID: 11302820.
35. White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997; 179:6122–6126. PMID: 9324261.
36. Yamane K, Wachino Ji, Suzuki S, Arakawa Y. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob Agents Chemother. 2008; 52:1564–1566. PMID: 18285488.
37. Yang H, Duan G, Zhu J, Zhang W, Xi Y, Fan Q. Prevalence and characterisation of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes among Shigella isolates from Henan, China, between 2001 and 2008. Int J Antimicrob Agents. 2013; 42:173–177. PMID: 23796894.
38. Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990; 34:1271–1272. PMID: 2168148.
39. Yue L, Jiang HX, Liao XP, Liu JH, Li SJ, Chen XY, Chen CX, Lü DH, Liu YH. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet Microbiol. 2008; 132:414–420. PMID: 18573620.
40. Zurfluh K, Abgottspon H, Hächler H, Nüesch-Inderbinen M, Stephan R. Quinolone resistance mechanisms among extended-spectrum beta-lactamase (ESBL) producing Escherichia coli isolated from rivers and lakes in Switzerland. PLoS One. 2014; 9:e95864. PMID: 24755830.
Fig. 1
Correlations between organic solvent tolerance (OST) or number of target mutations and minimum inhibitory concentrations (MICs) of nalidixic acid (NA) and ciprofloxacin (CIP) among 27 NA-resistant E. coli isolates. The size of the closed circle in each dot plot represents the number of NA- or CIP-resistant E. coli isolates. The scale box located on the right side of graphs shows three different-sized closed circles with the corresponding number of NA- or CIP-resistant isolates. The gradient of the trend line in each dot plot represents positive or negative correlation between the two variables. R: correlation coefficient.
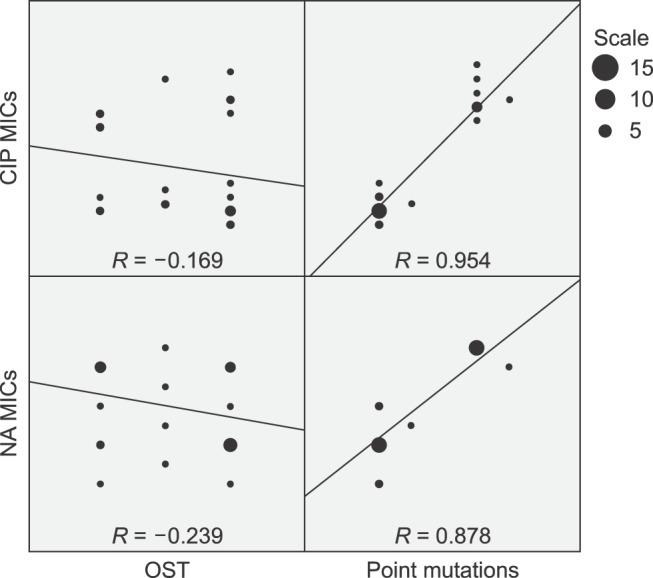
Table 1
Prevalence of nalidixic acid (NA)- or ciprofloxacin (CIP)-resistant Escherichia (E.) coli isolates from anal samples
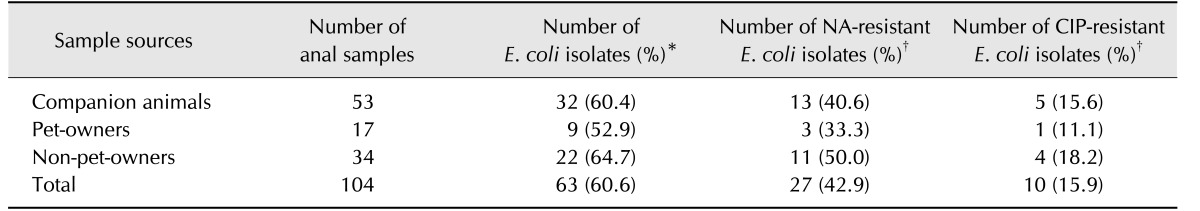
Table 2
Additional antimicrobial resistance profiling of 27 nalidixic acid (NA)-resistant Escherichia (E.) coli isolates
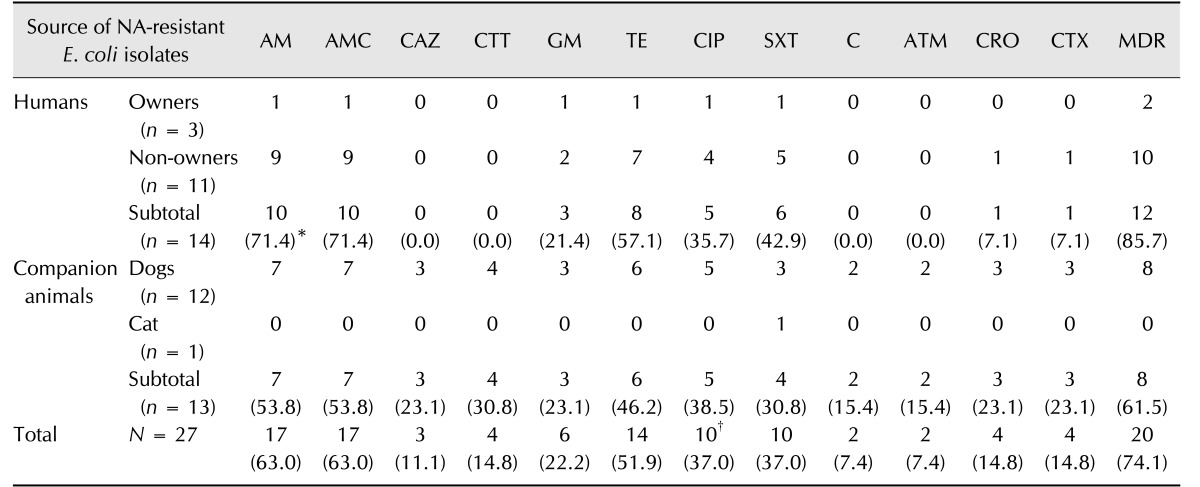
*The frequency of E. coli isolates showing resistance to each used antimicrobial is shown in the parenthesis. †All 10 CIP-resistant isolates were determined as multi-drug resistances (MDRs). AM, ampicillin; AMC, amoxicillin/clavulanic acid; CAZ, ceftazidime; CTT, cefotetan; GM, gentamicin; TE, tetracycline; CIP, ciprofloxacin; SXT, sulfamethoxazole/trimethoprim; C, chloramphenicol; ATM, aztreonam; CRO, ceftriaxone; CTX, cefotaxime.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download