Introduction
Since the discovery of the enteric protozoan parasite Cryptosporidium in mice by Tyzzer [37], Cryptosporidium infections have been detected in a wide range of vertebrate hosts throughout the world including more than 170 animal species and humans [28]. Numerous case reports have described infections of the gastrointestinal and respiratory tract, and revealed that Cryptosporidium could cause a potentially severe infection in immunosuppressed and immuodeficient hosts [9,11,26,28]. At present, Cryptosporidium species have been found in over 90 countries and six continents, including China [7,8], and 20 Cryptosporidium species are recognized [14].
Traditionally, cattle have been considered to be a primary reservoir for human Cryptosporidium infections and play a role in transmitting this parasite between humans and animals [25]. Cryptosporidium has been implicated as the cause of numerous outbreaks of watery diarrhea associated with contaminated food or water supplies [11,18,28], and some cases of water-borne transmission have been linked to domestic livestock, especially cattle [18]. Moreover, cryptosporidiosis has directly caused economic losses due to cattle death and treatment, and indirectly via decreased milk production capacity and increased production cost [28]. Although a large number of studies have demonstrated the efficacy of passive immunotherapy or chemotherapeutic agents for treating cryptosporidiosis, no significant clinical benefit has been demonstrated [8,18,38].
Nowadays, there are more than 13 million of dairy cattle in China [39], this figure places China in the front ranks of cattle-producing countries. To date, little information regarding the epidemiology and phylogeny of Cryptosporidium species in cattle from China has been reported [7,8]. Consequently, this study was designed to (a) assess the prevalence of Cryptosporidium in cattle in Anhui province, Jiangsu province and Shanghai city of eastern China, and (b) to precisely identify the species and genotypes of Cryptosporidium using molecular techniques.
Materials and Methods
Samples
This research was performed on four farms in Anhui province, six farms in Jiangsu province, and four farms in Shanghai city in eastern China. The 14 farms were randomly selected and visited between August 2007 and December 2009. A total of 2,056 fecal samples from dairy cattle were collected directly using disposable gloves and immediately analyzed within 24 h after collection.
Detection of Cryptosporidium oocysts
Triplicate fecal smears were prepared from each sample, and stained using the modified acid-fast staining technique (MAFS). Briefly, fecal smears were treated with a carbol-fuchsin solution (1 g fuchsine, 10 mL 95% ethanol, 5 g phenol, 95 mL distilled water) for 3 min, destained with a 1% hydrochloric acid-alcohol (70%) solution for 2 min, washed with running water, and counterstained with a 1% methylen blue solution for 1 min. After a final wash with water, the smears were dried at room temperature and examined by light microscopy using a 40× objective lens to verify the presence of Cryptosporidium oocysts. Oocyst size, shape, and staining characteristics were recorded. The average numbers of oocysts in 20 visual fields at a magnification of 400 × were classified as follows: '+' for 1~5 oocysts, '++' for 6~10, '+++' for 11~15, '++++' for 16~20, '+++++' for >20, and '-' for none.
Purification and morphologic evaluation of Cryptosporidium oocysts
Positive fecal samples were stored at 4℃ in a 2.5% aqueous potassium dichromate solution (K2Cr2O7). Prior to purification, each sample was filtered through an 80-mesh sieve to remove large debris, and the filtrate was processed within 24 h of collection as follows: K2Cr2O7 was removed by three cycles of precipitation with phosphate buffered saline (PBS; 0.1 M, pH 7.4) and suspended in PBS. Oocysts were then isolated using the saturated sucrose floatation technique [33] and further purified by discontinuous sucrose gradient centrifugation [36]. Oocysts were counted with a hemocytometer (Great Apparatus, China) and the sizes of 50 oocysts were measured using digital image analysis software (Image-Pro Plus 5.1; Media Cybernetics, USA) and a DP12 digital camera (BX41 microscope; Olympus, Japan).
DNA extraction
Cryptosporidium oocysts were washed three times with PBS and suspended in 0.2 mL sterile distilled H2O prior to DNA extraction. Genomic DNA was extracted using a DNA extraction kit (ver. 3.0; TaKaRa, Japan). The supernatant containing DNA was eluted in 50 µL of AE buffer(Qiagen, USA) and stored at -20℃ before it was used for polymerase chain reaction (PCR) analysis. Genomic DNA was also extracted from a negative sample (no oocyst DNA present) as a negative control.
Small subunit ribosomal RNA (SSU rRNA) gene amplification
A two-step nested PCR protocol was used to amplify a ~430 bp fragment of the SSU rRNA gene using primers 5'-gtggcaatgacgggtaacgg-3' and 5'-caggacatctaagggcatca-3' for the primary PCR, and 5'-aagctcgtagttggatttctg-3' (CPB-DIAGF) and 5'-taaggtgctgaaggagtaagg-3' (CPB-DIAGR) for the secondary PCR [22]. The PCR was carried out in 25 µL volume containing 2.0 µL (10 ng/µL) of genomic DNA, 2.5 µL 10× PCR buffer, 1.0 µL MgCl2 (50 mM), 0.2 µL (5 IU/µL) Taq DNA polymerase, 0.5 µL dNTP (10 mM), 1.0 µL (20 pmol/µL) of each forward and reverse primer, and nuclease free water up to 25 µL. The first step PCR program included 35 cycles (94℃ for 45 sec, 55℃ for 30 sec and 72℃ for 1 min), and the second PCR step included 40 cycles (94℃ for 30 sec, 55℃ for 30 sec and 72℃ for 1 min) using the DNA product of the first step PCR as the template.
Sequence and phylogenetic analysis
PCR products were analyzed by electrophoresis in a 1% (w/v) agarose gel using Tris-acetate-EDTA buffer (0.04 M Tris-acetate, 1 mM EDTA), purified using an agarose gel DNA purification kit (ver. 2.0; TaKaRa, Japan), and sequenced by TaKaRa (Japan). Sequence accuracy was confirmed by aligning the overlapping sequences obtained with an ABI 377 Automated Sequencer (Applied Biosystems, USA). Sequences from both orientations were aligned with reference sequences down-loaded from GenBank using the program Clustal V sequence alignment programs (DNAStar, USA).
The nucleotide sequences of the SSU rRNA gene fragment from Cryptosporidium were deposited into the GenBank database and a search for highly similar sequences was performed using nucleotide blast (NCBI, USA) to confirm the species or genotype. Phylogenetic trees were constructed from the SSU rRNA using the SeqEd and Cluster V sequence alignment programs (DNAStar, USA). In an initial neighbor-joining analysis, Eimeria tenella strain Wu (DQ136184) was used as an out-group. For comparative phylogenetic analysis, other sequences were retrieved from GenBank, including Cryptosporidium (C.) hominis strain H7 (AF108865), C. hominis strain HFL5: AF093492), C. bovis strain C1 (AF108864), C. bovis strain BOH6 (AF093490), C. parvum monkey strain CPRM1 (AF112569), C. parvum mouse M24 (AF108863), C. parvum strain RPHN (DQ898158), C. parvum strain CPM1 (AF112571), C. ryanae 2040 (AY120910), C. parvum ferret strain CPF (AF112572), C. parvum koala strain K2 (AF112570), C. parvum rabbit (AY120901), C. suis (AF108861), C. wrairi strain CWR (AF115378), C. meleagridis strain CMEL (AF112574), C. felis (AF108862), C. canis strain CPD1 (AF112576), C. baileyi genotype CBA01 (AF093495), C. andersoni strain bjcm (AY954885), C. andersoni IDVS-811 (AF093496), C. serpentis strain CSP01 (AF093499), C. serpentis isolate 119 (EU553560), C. saurophilum CSP06 (AF112573), and C. muris IDH-13 (AF093498), C. fragile clone A (EU162751).
Data analysis
For statistical evaluation of the results, SPSS 13.0 (SPSS, USA) and Microsoft Office Excel 2003 (Microsoft, USA) were used. Prevalence of Cryptosporidium spp. in different groups of cattle was compared with a Chi-square test for independence (α = 0.05). A correlation coefficient (r) was obtained and the significance of correlation between oocyst positivity and diarrhea was analyzed by Student's t-test. p-values < 0.05 were considered statistically significant.
Results
Epidemiology analysis
A total of 387 out of 2,056 feces samples were positive for Cryptosporidium as determined by MAFS, representing a prevalence rate of 18.82%. This included 69 of 350 samples from the Anhui province, 251 of 1,315 samples from the Jiangsu province, and 67 of 491 samples from Shanghai city, corresponding to the prevalence rates of 19.71%, 19.09% and 13.65%, respectively (Table 1). There was a significant difference in prevalence among these districts (χ2 = 8.07, p < 0.05). The prevalence in Anhui and Jiangsu provinces was higher than that in Shanghai city (χ2 = 5.55, 7.30, p < 0.05), but no significant difference in prevalence between the Jiangsu and Anhui provinces was observed (χ2 = 0.07, p > 0.05).
Among the four farms in the Anhui province, the prevalence rates were 20.69% (18/87), 21.30% (23/108), 18.07% (15/83), and 15.28% (11/72). No significant difference in prevalence among these farms was observed (χ2 = 1.21, p > 0.05). The prevalence rates among the six farms in the Jiangsu province were 20.70% (47/227), 20.00% (45/225), 23.08% (45/195), 17.80% (34/191), 19.75% (37/182), and 20.33% (43/195). Among the four farms in Shanghai city, the prevalence rates were 14.62% (19/130), 9.82% (11/112), 12.24% (12/98), and 16.56% (25/151). No significant difference in prevalence among these farms was observed (χ2 = 2.74, p > 0.05).
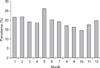
Prevalence of Cryptosporidium oocysts in the feces (Fig. 1) was highest in May (21.88%, 42/192) and lowest in October (14.29%, 22/154). There was no significant difference between the two months (χ2 = 3.26, p > 0.05), and no significant difference among the 12 months was observed (χ2 = 7.15, p > 0.05). The rates of infection in spring, summer, autumn, and winter were 19.42% (81/417), 19.17% (107/558), 16.16% (96/594), and 17.54% (103/587), respectively. There was no a significant difference in prevalence among the four seasons (χ2 = 2.54, p > 0.05).
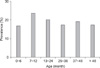
The prevalence of Cryptosporidium in cattle of different age ranged from 16.94% (41/242, 0~6 months old) to 23.81% (60/252, 7~12 months old) (Fig. 2). The youngest calf excreting oocysts was 6 days old and the oldest was 5 years old. Overall, the prevalence of Cryptosporidium infection did not vary significantly according to age (χ2 = 5.59, p > 0.05).
Out of the 387 positive samples, the intensity of infection ranged from '+' to '+++++', constituting 60.72%, 28.17%, 7.49%, 2.84%, and 0.78%, respectively. Cases of light infection, identified by scores of '+' and '++', represented 88.89% of the total.
The prevalence in cattle with and without diarrhea was also analyzed with a chi-square test. Out of the 245 samples from cattle with diarrhea and 1,811 samples from cattle without diarrhea, 81 and 306 were detected positive, respectively. This indicated a significant association between positivity and diarrhea (33.06% and 19.90%; χ2 = 36.90, p < 0.01).
Oocyst characteristics
Cryptosporidium oocysts, which were round or elliptic in shape, were observed by the contrast of their intense pink-rosy pigmentation against the yellow-green, blue, or glaucous background after staining with the MAFS technique. Cryptosporidium oocysts isolated by the saturated sucrose floatation technique and further purified by discontinuous sucrose gradient centrifugation showed different internal structures and cell size. One, two, or three sporozoites per oocyst were seen, but the total number in each oocyst could not be clearly visualized. Each oocyst also contained a residuum consisting of numerous small granules and a membrane-bound globule.
Cryptosporidium oocysts from different species varied in size and shape based on morphology and oocyst measurements. Oocysts (n = 50) from C. parvum 'mouse' measured 4.22~5.69 µm × 3.62~5.25 µm with a mean size of 4.98 µm × 4.46 µm, and had a shape index (the ratio of the length and width of oocysts) of 1.11. (1.05~1.32). Oocysts (n = 50) from C. andersoni measured 6.69~8.41 µm × 4.84~6.18 µm with a mean size of 7.46 µm × 5.55 µm, and a shape index of 1.34 (1.12~1.43). C. bovis oocysts (n = 50) measured 4.09~5.44 µm × 3.96~5.14 µm with a mean size of 4.83 µm × 4.64 µm, and a shape index of 1.06 (1.04~1.16). C. hominis oocysts (n = 50) measured 4.61~6.21 µm × 4.24~5.84 µm with a mean size of 5.18 µm × 4.98 µm, and had a shape index of 1.09 (1.02~1.18). Finally, oocysts (n = 50) from C. serpentis measured 5.73~6.49 µm × 4.96~5.38 µm with a mean size of 6.17 µm × 5.16 µm, and had a shape index of 1.18 (1.04~1.26).
Sequence and phylogenetic analysis of the SSU rRNA gene
Nucleotide sequences of the SSU rRNA gene fragment from Cryptosporidium were deposited into the GenBank database under accession numbers EU369377-84 and GU070730-33. Using the BLAST sequence analysis services of GenBank, we found that EU369377-80 was most similar to the sequences of C. andersoni IDVS-811 (AF093496), isolate 21 (FJ608605), and isolates (FJ463171-87) with 99% sequence identity. EU369381-84 were most similar to sequences of the C. parvum 'mouse' genotype strain M24 (AF108863), the RPHN strain (DQ898158), and CPM1 strain (AF112571) with 99~100% sequence identity. GU070730 was most similar to sequences of C. bovis strain C1 (AF108864), the BOH6 strain (AF093490), GCH1 strain (AF093493), DRI strain (AF093494), IOWA strain (AF164102); and strains AB513870-81, AB513857-68, and AF161856-59 with 100% sequence identity. GU070731 and GU070732 were most similar (100%) to sequences of C. hominis strain HFL5 (AF093492), and had a 99% identity with the B1b-B3b strain (AY204231-33), CPRM1 strain (AF112569), and Sakha isolates (AB513857-68, AB513870-80). GU070733 were most similar to the sequences of C. serpentis (AF151376) and C. serpentis CSP01 isolate (AF093499) with 100% identity, C. serpentis CSP02 isolate (AF093500) with 99.5% identity, and C. serpentis 40 isolate (EU553553) and the EU553558-83 isolate with 99% identity.
Phylogenetic (Fig. 3) and sequence homology analyses showed that EU369377-80 was placed in the C. andersoni branch with 97.9~100% homology, and adjacent to C. muris. EU369381-84 was placed in the C. parvum 'mouse' genotype branch with 98.4~100% homology, and adjacent to C. hominis and C. bovis. GU070730 was placed in the C. bovis branch with 100% homology, and adjacent to C. hominis and C. parvum. GU070731 and GU070732 were placed in the C. hominis branch with 98.2~100% homology, and adjacent to C. bovis, C. parvum 'rabbit' genotype, and C. parvum 'monkey' and 'mouse' genotype. GU070733 was placed in the C. serpentis branch with 100% homology, and adjacent to C. ryanae and C. baileyi.
No cattle were detected with a mixture of Cryptosporidium species or genotypes. Of the 387 PCR-positive specimens characterized by gene sequencing, 185, 112, 62, 24, and 4 were infected with C. parvum 'mouse' genotype, C. andersoni, C. bovis, C. hominis, and C. serpentis, respectively. The percentages of each species or genotype of Cryptosporidium relative to the total number of Cryptosporidium-positive cattle were 47.80%, 28.94%, 16.02%, 6.20%, and 1.02%, respectively.
Discussion
Cryptosporidium was first isolated in cattle from a case of calf diarrhea in 1971 [30]. Since then, there have been numerous reports of cryptosporidiosis in different countries and regions [5,6,17,19,20,27]. However, only a few publications have documented Cryptosporidium prevalence among animals in China [7,40]. Because cattle breeding in China is an important agricultural activity and given high potential for cattle-to-human transmission of Cryptosporidium [25], it is of interest to investigate Cryptosporidium epidemiology in these animals according to the set of the best means as described by other investigators [3,5,7,12,13]. In the present study, we systematically investigated the prevalence and genetic identification of Cryptosporidium species in dairy cattle in China. We found that 387 (18.82%) samples were positive for Cryptosporidium oocysts in 2,056 fecal samples of dairy cattle from 14 farms using MAFS. Phylogenetic and sequence analysis confirmed the existence of C. parvum 'mouse' genotype, C. bovis, C. andersoni, C. hominis, and C. serpentis in the fecal samples, and C. parvum 'mouse' and C. andersoni were the most general Cryptosporidium species. These results represent the first report on the prevalence and genetic identification of Cryptosporidium species in dairy cattle in China and may contribute to a better understanding of Cryptosporidium epidemiology in cattle.
In the current study, Cryptosporidium was not found only in specific farms or provinces (cities), but was present on all 14 farms in three districts of eastern China. The average rate of infection was 18.82%, and ranged from 9.82 to 23.08% on different farms. This level of infection was higher than that found in western Canada (1.1%) [17], Italy (4.99%) [10], Spain (8.4%) [6], Thailand (13% by the acid-fast staining technique and 9.63% by nested PCR) [27], Galicia (14.2%) [5], and Alberta (2.1% and 5.8%) [20]. The infection rate in our study was also similar to that reported in Canada (20%) [29] and Aragón (21.3%) [31], but was lower than that in California (28%) [21], Mongolia (26.4%) [4], and Malaysia (36%) [19]. Many factors may contribute to different prevalence rates, including the climate and farm practices in different countries. It would be interesting to investigate whether cattle-to-human or cattle-to-other animal transmission of Cryptosporidium is relevant to public health.
Quilez et al. [31] reported that the prevalence of cryptosporidiosis is generally lower in neonatal animals due to maternal protection. Scott et al. [33] found no relation between the presence of oocyst-excreting adults and the development of infection in calves. de la Fuente et al. [9] reported that the detection rate of Cryptosporidium in animals 22~30 days old was significantly lower than those 1~21 days old in central Spain. In contrast, the present study found that the prevalence of Cryptosporidium was not different among cattle of different ages, suggesting that adult cattle might act as reservoirs for Cryptosporidium that infects neonatal and younger cattle.
Many authors have reported contradictory findings regarding the seasonal prevalence of Cryptosporidium on cattle farms [23,34]. Mohammed et al. [23] found that dairy cattle sampled in spring in southeastern New York State were significantly less likely to be positive for Cryptosporidia compared to cattle sampled in the winter. In contrast, Becher et al. [2] reported that there was no difference in seasonal prevalence on cattle farms in Western Australia. In the present study, we found no seasonality in Cryptosporidium prevalence, indicating high levels of humidity which frequently occur in all seasons in the study regions may favor transmission.
Cryptosporidium is considered to be one of the causative enteropathogens of diarrhea in cattle [9], although asymptotic infection had also been described [26]. In the present study, the infection rate among cattle with diarrhea was higher than that among asymptomatic animals. Therefore, we concluded that cryptosporidiosis is an important cause of diarrhea in cattle.
Many studies have suggested that cattle are infected with at least four Cryptosporidium species (C. parvum, C. bovis, C. andersoni, and C. ryanae) in the United States [12,13,19,23,32]. Feng et al. [16] reported that C. bovis and Cryptosporidium deer-like genotype were found in all age groups of cattle from diverse geographic areas, and were much more prevalent in post-weaned cattle [12]. Fayer et al. [12,13] found that C. andersoni were found in more cows or cattle while C. bovis, C. parvum, and Cryptosporidium deer-like genotype were less prevalent [32]. Feltus et al. [15] reported that C. bovis, the deer-like genotype, and C. andersoni were identified in 9.4, 6.6, and 1.4% of the animals sampled, respectively. Zhou et al. [40] examined C. andersoni infection in dairy cattle in China. In this study, five Cryptosporidium species or genotypes were identified according to oocyst dimension and genetic analysis of SSU rRNA. C. parvum 'mouse', C. hominis, C. serpentis, and C. bovis were first found in cattle in China, and the infection rates of C. parvum 'mouse' and C. andersoni were higher than those of other genotypes. C. andersoni and C. bovis were not the most general Cryptosporidium found in cattle. This finding was similar to other investigations showing that the infection rate of C. andersoni was lower among cattle [32], but was different from studies conducted in England and northern Ireland [3,36]. Based on these findings, it is possible to conclude that geographic condition, climate, feeding, and management may affect the prevalence of Cryptosporidium in cattle.
Morgan et al. [24] reported that C. parvum 'mouse' genotype was primarily isolated from wild rats and house mice, and wild rodents were thought to serve as an important reservoir. In the present study, C. parvum 'mouse' genotype was the most prevalent species in the 387 positive samples, constituted 47.8% of the positive samples. When sampling, we found that there were some mice and mouse feces in the farms, but we could not collect and detect those samples. Whether cryptosporidiosis is transmitted between rodents and cattle in eastern China remains to be investigated.
Morgan-Ryan et al. [25] reported that C. hominis was non-infectious in mice, rats, cats, dogs, and cattle, but infectious in calves, lambs, and piglets. C. serpentis was originally described as a gastric parasite found in lizards and other reptiles, but there are few reports of C. serpentis infection in other animals or humans [19]. Only Azami et al. [1] reported that C. serpentis can infect cattle. Similarly, C. hominis and C. serpentis infections were found in 6.20% and 1.02% of the cattle in the present study. Oocysts are transmitted from an infected host to a susceptible entity by the fecal-oral route, either through direct animal-to-human contact or water-borne routes via contaminated drinking water [11]. Although we did not confirm the transmission routes of C. hominis and C. serpentis, it is of great important to enhance the management of cattle and drinking water and to reduce transmission between cattle and humans or other animals and their feces in order to cut off the route of Cryptosporidium transmission to cattle.
In conclusion, there was a high prevalence of Cryptosporidium in dairy cattle on farms in China. Cryptosporidium species infecting the cattle in China belonged to C. parvum 'mouse' genotype, C. andersoni, C. bovis, C. hominis, and C. serpentis, and C. parvum 'mouse' and C. andersoni were the most general Cryptosporidium species.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download