Abstract
It is well established that blocking the interaction of EGFR with growth factors leads to the arrest of tumor growth, resulting in tumor cell death. ER414 is a human monoclonal antibody (mAb) derived by guided selection of the mouse mAb A13. The ER414 exhibited a ~17-fold lower affinity and, as a result, lower efficacy of inhibition of the EGF-mediated tyrosine phosphorylation of EGFR when compared with mAb A13 and cetuximab. We performed a stepwise in vitro affinity maturation to improve the affinity of ER414. We obtained a 3D model of ER414 to identify the amino acids in the CDRs that needed to be mutated. Clones were selected from the phage library with randomized amino acids in the CDRs and substitution of amino acids in the HCDR3 and LCDR1 of ER414 led to improved affinity. A clone, H3-14, with a ~20-fold increased affinity, was selected from the HCDR3 randomized library. Then three clones, ER2, ER78 and ER79, were selected from the LCDR1 randomized library based on the H3-14 but did not show further increased affinities compared to that of H3-14. Of the three, ER2 was chosen for further characterization due to its better expression than others. We successfully performed affinity maturation of ER414 and obtained antibodies with a similar affinity as cetuximab. And antibody from an affinity maturation inhibits the EGF-mediated tyrosine phosphorylation of EGFR in a manner similar to cetuximab.
Epidermal growth factor receptor (EGFR) is a 170-kDa membrane-spanning glycoprotein comprising an extracellular ligand-binding domain, a transmembrane domain, and an intracellular cytoplasmic domain that has tyrosine kinase activity (1). EGFR belongs to the human epidermal receptor (HER) family of receptor tyrosine kinases, which consists of four closely related receptors-EGFR (HER1, erbB1), HER2 (neu, erbB2), HER3 (erbB3), and HER4 (erbB4)-that mediate cellular signaling pathways involved in growth and proliferation in response to the binding of a variety of growth factor ligands (2,3).
Activation of EGFR has been shown to enhance processes responsible for tumor growth and progression, including proliferation, angiogenesis, invasion and metastasis, and inhibition of apoptosis. Expression of EGFR varies widely in tumors, including head and neck (80~100%), renal (50~90%), lung (40~80%), breast (14~90%), colorectal (25~77%), ovarian (25~70%), prostate (39~47%), glioma (40~63%), pancreas (30~50%) and bladder (31~48%) (4,5). High levels of EGFR protein expression in tumors have been correlated with aggressive disease, poor prognosis and decreased survival, poor response to therapy and the development of resistance to cytotoxic agents in some tumor types (5,6).
The clear potential for EGFR-targeted therapies in the treatment of cancer has prompted the development of a variety of agents targeted to the extracellular ligand-binding domain, the intracellular tyrosine kinase domain, the ligand, or the synthesis of EGFR (7,8). Among these, the two most extensively studied therapies to date consist of monoclonal antibodies (mAbs) directed against the extracellular receptor domain and small-molecule compounds that interfere with intracellular EGFR tyrosine kinase activity (9).
Previously, we reported the generation of ER414, a human mAb to EGFR, by guided selection of the well characterized murine mAb A13 (10). The ER414 exhibited a ~17-fold lower affinity compared to mAb A13 and inhibited EGF-mediated EGFR tyrosine phosphorylation in breast tumor cells with lower efficacy than mAb A13 and cetuximab (10).
In this article, we report the affinity maturation of ER414. For this purpose, phage libraries with randomized amino acids in the HCDR3 were constructed, and clones with increased affinity were selected by panning against EGFR. Next, phage libraries with randomized amino acids in the LCDR1 were constructed from the clone, which contained HCDR3 mutations that resulted in an increased affinity, and the clones were selected by panning against EGFR. Finally, clones from the HCDR3 and LCDR1 mutations were compared with cetuximab for their affinity and ability to inhibit EGF-induced tyrosine phosphorylation of EFGR. In addition, the binding site of one of the mAb that showed increased affinity was analyzed using yeast surface-displayed EGFR fragments and Surface Plasmon Resonance (SPR).
The 3D structure of ER414 was predicted by the Web Antibody Modeling (WAM; http://antibody.bath.ac.uk) software. Amino acids are numbered sequentially. In the model, VH is shown in brown and VL is shown in cyan. HCDR3 (green) and LCDR1 (red) appear to contribute mainly to antigen binding and are highlighted (Fig. 1).
Random sequences were introduced at positions Ser102, Gly103, Val105, Asp106, Gly 108 and Met109 in the HCDR3 of ER414 by PCR via simultaneous saturation mutagenesis using degenerate oligonucleotides NNS (N=A/C/G/T and S=C/G). DNAs were obtained using the two primer pairs, H-extension(F)/H-12(R) and H-11(F)/H-extension(R) in a PCR. These two DNA fragments were assembled in a PCR using the set of primers H-extension(F)/H-extension(R), which contained the Sfi I and BstE II restriction sites, respectively. The primer sequences are summarized in the Table I.
Another set of random mutations were introduced at positions Gly100, Ser101, Ser102, Gly103, Val105 and Asp106 in the HCDR3 of ER414 in a PCR amplification of DNAs using the primers H-extension(F)/H-14(R) and H-13(F)/H-extension(R). Subsequently, the two DNA fragments were assembled using the set of primers H-extension(F)/H-extension(R) in a PCR (Table I). The final PCR product was digested with enzymes Sfi I and BstE II (New England BioLabs, Ipswich, MA), and the digestion products were inserted into the plasmid pSC73-ER414 (Fig. 2); the resulting plasmids were designated as pSC73-ER414-rH3A and pSC73-ER414-rH3B. Each of the plasmids, pSC73-ER414-rH3A and pSC73-ER414-rH3B, was introduced into E. coli XL1-Blue by electroporation, and phage particles that displayed ER414 with HCDR3 randomization were produced by the addition of M13 helper phages.
H3-14 is an affinity-improved ER414 obtained by introducing mutations in the HCDR3 and was used as a template for obtaining new clones upon LCDR1 randomization.
Random sequences were introduced at positions Ser26, Ser28, Leu29, Leu30, His31, Ser32, Asn33, and Asn36 in the LCDR1 of H3-14. DNAs were obtained in a PCR using the two primer pairs K-extension(F)/K-11(R) and K-10(F)/K-extension(R). Subsequently, the two DNA fragments were assembled in a PCR using the set of primers K-extension(F)/K-extension(R), which contained the BstE II and Not I restriction sites, respectively (Table I).
Another set of random sequences was introduced at positions Leu29, Leu30, His31, Ser32, and Asn33 in the LCDR1 of H3-14 in a PCR amplification of the DNAs using the two primer pairs K-extension(F)/K-13(R) and K-12(F)/K-extension(R), followed by the assembly of the two DNA fragments in another PCR utilizing the set of primers K-extension(F)/K-extension(R) (Table I). The final PCR product was digested with enzymes BstE II and Not I (New England BioLabs) and inserted into the plasmid pSC73-H3-14; the resulting plasmids were designated as pSC73-rL1A and pSC73-rL1B. Each of the plasmids, pSC73-rL1A and pSC73-rL1B, was introduced into E. coli XL1-Blue by electroporation, and phage particles that displayed H3-14 with LCDR1 randomization were produced by addition of M13 helper phages.
Phages binding to EGFR were selected by panning in an immunotube (NUNC, Roskilde, Denmark) as described earlier (10). Tubes were coated with 1 ml of 5 µg/ml EGFR (Sigma, St. Louis, MO) in PBS and blocked with 1% BSA-PBS. The phages displaying the ER414 with randomized CDRs were incubated for 2 hr at 37℃ in an EGFR-coated tube, and the tube was washed with PBS-T (PBS buffer containing 0.05% Tween 20). The phages were eluted from the tube with 0.1 M Glycine buffer (pH 2.0) containing 1% BSA and neutralized with 2 M Tris. The E. coli XL1-Blue cells were infected with the neutralized phages, followed by an infection with M13 helper phages, and grown overnight at 37℃. The phages were isolated by PEG precipitation and were used for the next round of panning.
Expression of scFv was performed in colonies that were obtained after the fifth round of panning, as described above (10). Five milliliters of SB containing 50 µg/ml of carbenicillin were inoculated with a colony and grown at 37℃ with continuous shaking until the O.D600 reached a value of 1. IPTG (isopropyl-β-D-thiogalactopyranoside, Sigma), at a concentration of 1 mM, was added, and the bacteria were allowed to grow overnight at 30℃ with continuous shaking. The culture was centrifuged, and the supernatant was analyzed for the expression of scFv.
ELISA was performed to measure the binding of the expressed scFv to EGFR, as described (11-13). Each well of the ELISA plates (NUNC Immuno Module, Maxisorp) was coated with 100 µl of 2 µg/ml EGFR in PBS and incubated overnight at 4℃ followed by blocking with 300 µl of 1% BSA-PBS for an hour at room temperature. A 100-µl aliquot of the supernatant from the bacterial culture was added to the plate and incubated for 2 hr at room temperature. After washing with PBS-T (PBS buffer containing 0.05% Tween 20), 100 µl of diluted HRP/Anti-His Tag Conjugate (Qiagen, Valencia, CA) in 1% BSA-PBS was added, and the plate was incubated for 1 hr at room temperature. After a second wash with PBS-T, 100 µl of TMB 2-component microwell peroxidase substrate (KPL, Gaithersburg, MD) was added, and the O.D. was measured at 405 nm.
Sequencing of immunoglobulin genes was performed in Genotech (Daejeon, Korea) using the primer Seq 001 (Table I), and sequence analyses were performed using the CLC main workbench (CLC bio, Cambridge, MA).
VH and VL selected from panning were inserted into the mammalian expression vectors pRC-12 (Fig. 3) for the heavy chain and pKC-12 (Fig. 3) for the light chain, respectively, as previously described (14). The heavy and light chain expression vectors were co-transfected into CHO DG44 cells (15) using Effectene (Qiagen), and all of the procedures for the screening and selection of cells were followed as described previously (14). Cells producing anti-EGFR antibody were screened by ELISA as described previously (16).
Antibodies were purified using protein A from the culture media. Antibodies affinity was compared by competition ELISA on EGFR-coated plates in the presence of free EGFR, as described (11,14). The EGFR concentration that gives 50% inhibition of the maximum binding (the ELISA reading performed without competitive EGFR) was considered to be the affinity.
The binding of selected scFvs to cell surface-expressed EGFR was analyzed in A431 cells by FACS (FACScan, Becton-Dickinson, Mountain View, CA). Briefly, 1×107 cells were harvested and washed with 1% BSA-PBS and incubated with 10 µg/ml of mAb A13, ER414 or ER2 for 1 hr on ice. After being washed with 1% BSA-PBS, the cells were incubated with FITC-conjugated goat anti-human IgG (Fab specific) (Sigma) for ER414 and ER2 or with FITC-conjugated goat anti-mouse IgG (Fab specific) (Sigma) for mAb A13 in 1% BSA-PBS for a period of 40 min on ice and then analyzed by FACS.
To analyze the inhibition of EGFR tyrosine phosphorylation by mAbs, 1×105 of MDA-MB-231 cells were plated in 24-well culture plates (Nunc) and serum-starved for 48 hr before EGF and the antibody were added. After EGF treatment (16 nM) with different amounts of antibodies (from 33.3 pM to 666.7 pM) for 30 min, cells were washed and lysed in TritonX lysis buffer (17) (10 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 2 µg/ml leupeptin & aprotinin, 1 mM Na3VO4, 1 mM PMSF). An equal amount of protein (100 µg/ml) was boiled with the sample buffer for 5 min and run on a 10% SDS-PAGE gel. The separated proteins were transferred onto a nitrocellulose membrane (BIO-RAD, Hercules, CA) and probed with mouse anti-phosphotyrosine-HRP (Zymed) and 4CN 2-Component Membrane Peroxidase Substrate Kit (KPL).
The pCTCON yeast display plasmids containing the appropriate genes that encode for the EGFR fragments were used for domain-level epitope mapping of ER2, as previously reported (16). Each plasmid was transformed into the yeast strain EBY100 by electroporation using a Bio-Rad Gene Pulser electroporation (Richmond, CA) apparatus (18,19). The transformant propagation and induction for the cell surface expression of the EGFR fragments were performed as described previously (18,19). The cell surface expression level of the EGFR fragments and the mAb binding to the cell surface-expressed EGFR fragments were determined using flow cytometry by indirect single immunofluorescence labeling, as previously described (18,19). The induced yeast cells (~5×106 cells) were labeled with either mouse anti c-myc 9E10 (Ig Therapy, Korea) or ER2 and then incubated with either FITC-conjugated anti-mouse IgG (Sigma) or FITC-conjugated anti-human IgG (Sigma).
The ER2 and cetuximab binding sites on EGFR were compared using the Biacore 2000 SPR biosensor (BIAcore AB, Uppsala, Sweden) as described previously (19,20). The antigen sEGFR was first immobilized on the CM5 chip at a level of ~4,000 response units, and 60 µl of cetuximab (500 nM) was injected into the flow cell at a flow rate of 30 µl/min for 120s. ER2 (500 nM) was immediately injected at the same flow rate for 180s before the dissociation phase.
We performed affinity maturation of ER414 initially by randomization of the HCDR3 and identified 2 clones, H3-14 and H3-15, from the HCDR3 randomized library, and their affinities were compared to that of ER414 by competition ELISA. In H3-14, Ser102, Val105, Asp106, Gly108 and Met109 were changed to Pro, Thr, Leu, Ala and Trp, respectively; the affinity of H3-14 showed a ~20-fold increase (Table II & Fig. 4). In H3-15, Gly100, Ser101, Ser102, Gly103 and Asp106 were changed to Ser, Trp, Gly, Ala and Gln, respectively; the affinity of H3-15 showed a 4.5-fold increase (Table II & Fig. 4).
Then, we performed randomization of the LCDR1 using H3-14 as the template. Three clones, ER2, ER78 and ER79, were identified from the LCDR1 randomized library. In ER2, Ser26, Ser28, Leu30 and Asn36 were changed to Asn, Asp, Thr and Thr, respectively (Table III). In ER78, Leu29, Leu30, His31, Ser32 and Asn33 were changed to Met, Val, Asp, Glu and Tyr, respectively (Table III). In ER79, Leu29, Leu30, His31, Ser32 and Asn33 were changed to Val, Asp, Met, Gly and Ile, respectively (Table III). The affinities of three clones were compared to that of H3-14 by competition ELISA; none of the three clones exhibited any increase in affinity when compared to H3-14 (Table III & Fig. 5).
Thus, ER2, ER78 and ER79 demonstrated a ~20-fold increase in affinity when compared to the wild type ER414, and this affinity is similar to that of cetuximab (Fig. 5).
Binding of ER2 to EGFR in tumor cells was examined by FACS analysis with A431 tumor cells along with mAb A13 and ER414. A431 tumor cells are known to express EGFR at high levels. All three antibodies, mAb A13, ER414 and ER2, showed considerable binding to the cell surface-expressed EGFR (Fig. 6).
The abilities of mAb A13, ER414, ER2 and cetuximab to block the EGF-mediated tyrosine phosphorylation of EGFR were compared using MDA-MB-231 cells. Tyrosine phosphorylation of EGFR was induced in the presence of 16 nM EGF (Fig. 7). There was a basal level of tyrosine phosphorylation of EGFR even when no EGF was present (Fig. 7). ER2 and cetuximab inhibited the EGF-induced tyrosine phosphorylation of EGFR with similar efficacy, of up to 33.3 pM antibody, whereas ER414 could not inhibit the EGF-induced tyrosine phosphorylation of EGFR at this concentration (Fig. 7).
The extracellular domain of EGFR can be divided into four subdomains: I (residues 1~165), II (residues 166~310), III (residues 311~480), and IV (residues 481~621) (18,21,22). To determine the domain level epitope of ER2 mAb on EGFR, the fragments corresponding to the extracellular domains of EGFR were expressed on the surface of yeast cells (16) and tested for ER2 mAb binding using flow cytometry by indirect immunofluorescence. For the whole EGFR, an engineered EGFR with four mutations at A62T, L69H, F380S, and S418G, designated as 404SG, was used instead of the wild-type EGFR because the 404SG mutant expressed well functionally and was recognized by many EGFR-specific mAbs on the yeast surface (18). EGFR fragments 1~124 (domain I), 1~176 (domain I), 1~294 (domain I+II), 166~503 (domain II+III), 273~621 (domain III+IV), 302~503 (domain III), 294~543 (domain III), and 475~621 (domain IV) were expressed at different levels on the yeast cell surface, as inferred from significant positive labeling by the anti-c-myc mAb (Fig. 8). The EGFR fragment 475~621 was hardly expressed, probably due to either improper folding or trafficking via the yeast secretory pathway (18,21). As shown in Fig. 8, ER2 mAb strongly interacted with the EGFR fragments 166~503, 273~621, 302~503, and 294~543, each of which contained domain III. The ER2 mAb did not bind to the EGFR fragments 1~124, 1~176, and 1~294, which contained domain I and/or II, despite their significant expression on the yeast cell surface. The binding of ER2 to the EGFR fragment 475~621, which contained domain IV, was negligible, but the lower fluorescence signal could have resulted from the fact that the fragment containing domain IV was not well expressed on the yeast cell surface (Fig. 8) (21). It can therefore be concluded that the ER2 mAb epitopes are most likely contained within the amino acid sequence 302~503, namely domain III of EGFR.
The binding sites of ER2 and cetuximab on EGFR were compared by competitive SPR analysis. In this analysis, ER2 was found to bind to the surface-coated sEGFR even after its saturated binding to cetuximab (Fig. 9). This result indicates that ER2 and cetuximab bind to EGFR at two distinct binding regions.
These observations suggest that ER2 binds to domain III of EGFR but at distinct epitopes that are not shared by cetuximab.
This study involved the antibody engineering of a mouse mAb A13 that targets the EGFR by antibody selection via guided panning and affinity maturation.
mAb A13 was generated from mice that were immunized with human cervical carcinoma A431 cells. mAb A13 specifically binds to a variety of tumor cells and human placenta tissues expressing the EGFR and efficiently inhibits both EGF-mediated EGFR tyrosine phosphorylation in cervical and breast tumor cells and the in vitro colony formation of EGFR-overexpressing lung tumor cells (16). Competition and sandwich ELISAs, competitive surface plasmon resonance, and domain-level epitope mapping analyses demonstrated that mAb A13 competitively bound to domain III (amino acids 302~503) of the EGFR in the presence of EGF but recognized distinct epitopes from those recognized by cetuximab (16).
ER414 is a human anti-EGFR IgG1 mAb derived from the mouse mAb A13 by guided selection (10). It was isolated from the hybrid scFv libraries containing a human VH repertoire with the VL of mAb A13 and a human VL repertoire with the VH of mAb A13. The ER414 exhibited a ~17-fold lower affinity than mAb A13 and cetuximab; ER414 inhibited the EGF-induced tyrosine phosphorylation of EGFR with a much lower efficacy compared to mAb A13 and cetuximab (10). The affinity of ER414 needed to be improved for clinical applications.
We performed a stepwise in vitro affinity maturation procedure to improve the affinity of ER414. We obtained a 3D model of ER414 to identify the amino acids in the CDRs that needed to be mutated. In the 3-D model of ER414, the HCDR3 and LCDR1 seemed to be involved in EGFR binding to a greater extent than other CDRs (Fig. 1). The heavy chain is considered to contribute to antigen binding to a greater extent than the light chain, especially through the HCDR3 (23); therefore, mutagenesis in the HCDR3 was attempted first. We identified 2 clones; H3-14, which had a ~20-fold increased affinity, and H3-15, which had a ~4-fold increased affinity. In the 3-D model of ER414, Tyr104, which is at the apex of the HCDR3 (Fig. 1), is likely crucial for binding to EGFR and was, therefore, not mutated. The differences in the affinities of H-14 and H-15 might have been caused by the changes around Tyr-104. The mutations introduced in H3-14 might have contributed to better positioning of the Tyr104 residue for tighter binding to EGFR than did the mutations in H3-15. The mutations introduced in H3-15 were farther away from the Tyr-104 than were those of H3-14. In the case of cetuximab, Tyr104 in the HCDR3 (amino acids are numbered sequentially) is at the center of the interface between Fab C225 and sEGFR and protrudes into a hydrophobic pocket on the surface of the large β sheet of domain III (24).
Then, we identified 3 clones (ER2, ER78 and ER79) that did not exhibit any increased affinity following the mutagenesis in the LCDR1 of H3-14. We assumed that the LCDR1 of H3-14 (or ER414) might not be involved in binding to the EGFR, although in the model, LCDR1 seems to be involved in the binding, as does HCDR3. Finally, we selected ER2 for further characterization because it could be produced in larger quantities compared to others (data not shown).
We adjusted the affinity of ER2 to be similar to that of cetuximab because increased affinity did not necessarily enhance tumor uptake. Antibodies with extremely high affinity have impaired tumor penetration properties; as the affinity of the antibody for its target antigen increases, the distribution becomes more perivascular (25,26).
Cetuximab binds to a site on domain III of the EGFR that overlaps with the EGF binding site, thereby blocking the binding of the ligands, EGF and TGF-α, to EGFR and subsequently preventing EGFR dimerization and activation (24,27). Previous studies have shown that the anti-EGFR mAbs, cetuximab and 425 (27), as well as cetuximab and matuzumab (Fab72000) (28), did not compete with each other in spite of their exclusive binding to domain III of the EGFR.
Epitopes of ER2 and cetuximab reside in domain III of EGFR but are different from each other, as confirmed by the domain-level epitope mapping of ER2 using yeast surface-displayed EGFR fragments and by Surface Plasmon Resonance (SPR). The binding of ER2 to the EGFR fragments that contain only domain III (fragments, 294~543 and 302~503) implies that it is feasible for ER2 to bind to the wild-type EGFR and to the truncated variant EGFRvIII (de2-7 EGFR), in which amino acids 6~273 have been deleted and which is also found to be frequently overexpressed in glioma (22).
Treatments that use a combination of mAbs with different binding epitopes on the same antigen have shown significant synergistic effects in tumor therapy. For example, co-treatments of anti-EGFR mAbs with distinct epitopes (e.g., cetuximab and matuzumab, or cetuximab and mAb 806) accelerated the death of breast cancer cells more effectively than when either mAb was used alone (22,27). Because ER2 and cetuximab have distinct epitopes, it may be possible that the combination of ER2 and cetuximab is a more effective strategy to target tumors that contain overexpressed EGFRs.
Cetuximab, which is approved for use in advanced colorectal cancer and in head and neck squamous-cell carcinoma, is a chimeric mAb and elicits immune reactions in ~19% of cases (29). In contrast, IMC-11F8, a human mAb that has antitumor potency that is similar to cetuximab, has shown no evidence of immune hypersensitivity in clinical trials (29). Likewise, ER2 is a human mAb and expected of no immune hypersensitivity although it is proved through clinical trials.
Figures and Tables
Figure 1
3D model of ER414 using the program Web Antibody Modeling (WAM; http://antibody.bath.ac.uk). Amino acids are numbered sequentially. VH is in brown, and VL is in cyan; HCDR3 (green) and LCDR1 (red) are highlighted.

Figure 2
The arrangement of genes in the pSC73 vector. LacZ denotes the lac promoter. PelB denotes the leader peptide of pectate lyase B of Erwinia carotovora, and GeneIII denotes gene3, which encodes the minor coat protein of the filamentous phage M13. His denotes a tag of 6 histidine repeats. Restriction enzyme sites used for cloning of scFv are shown.

Figure 3
The arrangement of genes in the expression vectors for the heavy (pRC12) and light (pKC12) chains. PCMV denotes cytomegalovirus promoter, and Sig denotes signal peptide of human immunoglobulin. Restriction enzyme sites used for cloning are shown. The light chain expression vector contains the dihydrofolate reductase (dhfr) gene (not shown) used for amplification of integrated genes by adjusting the cells in increasing concentrations of methotrexate (MTX).
Figure 4
Competition ELISA for measuring relative affinities of anti-EGFR IgGs from random mutations in the HCDR3 of ER414. The inhibition of anti-EGFR IgG binding to EGFR was analyzed on EGFR-coated plates with different concentrations of free EGFR, and the curves were fitted to a 4-parameter model using SoftMaxPro software. The antibodies are H3-14 (▴), H3-15 (♦) and ER414 (•).

Figure 5
Competition ELISA for measuring the relative affinities of anti-EGFR IgGs from random mutations in the LCDR1 of H3-14. The inhibition of anti-EGFR IgG binding to EGFR was analyzed on EGFR-coated plates with different concentrations of free EGFR, and the curves were fitted to a 4-parameter model using the SoftMaxPro software. The antibodies are ER2 (▪), ER78 (♦), ER79 (▴), ER414 (•) and cetuximab (○).

Figure 6
Binding of anti-EGFR mAbs to cell surface-expressed EGFR. Differential binding of mAbs to EGFR-positive A431 tumor cells was determined by flow cytometry. Histograms in different colors represent the reactions of labeled secondary Ab with A431 cells (black), ER414 with A431 cells (green), ER2 with A431 cells (blue), and mAb A13 with A431 cells (red).

Figure 7
Blocking of the EGF-induced tyrosine phosphorylation of EGFR by A13 mAb, ER414, ER2 and cetuximab in MDA-MB-231 cells. The serum-starved cells were untreated, treated with EGF (16 nM) only, or treated with EGF (16 nM) and 33.3, 166.7, 333.3 or 666.7 pM of mAbs for 30 min, as indicated in the panels, before the Western blotting analyses. The arrows indicate the expected size (~170 kDa) of the EGFR.
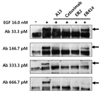
Figure 8
Domain-level epitope mapping of ER2 using the yeast surface-expressed EGFR fragments. Representative flow cytometry histograms depict anti-c-myc 9e10 labeling for the expression of EGFR fragments (black) and ER2 binding (blue) to the yeast surface-expressed EGFR fragments, as indicated in each panel. For the whole EGFR (1~621), the 404SG mutant was used (described in detail in the text).

Figure 9
Comparison of the ER2 and cetuximab binding sites on EGFR by competitive SPR assay. The first part of the curve shows the response for the binding of cetuximab (500 nM) to the surface-immobilized sEGFR, and the second part of the curve shows the binding upon injection of ER2 (500 nM) before the dissociation phase.

ACKNOWLEDGEMENTS
This work was supported by a grant from the Korea Biotech R&D Group's Next-generation of growth engine project through the Ministry of Education, Science and Technology, Republic of Korea (grant number: 2010K001288), and the Green Cross Corp., Korea.
References
1. Carpenter G. Receptors for epidermal growth factor and other polypeptide mitogens. Annu Rev Biochem. 1987. 56:881–914.

2. Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001. 2:127–137.

3. Laskin JJ, Sandler AB. Epidermal growth factor receptor: a promising target in solid tumours. Cancer Treat Rev. 2004. 30:1–17.

4. Herbst RS, Shin DM. Monoclonal antibodies to target epidermal growth factor receptor-positive tumors: a new paradigm for cancer therapy. Cancer. 2002. 94:1593–1611.

5. Capdevila J, Elez E, Macarulla T, Ramos FJ, Ruiz-Echarri M, Tabernero J. Anti-epidermal growth factor receptor monoclonal antibodies in cancer treatment. Cancer Treat Rev. 2009. 35:354–363.

6. Brabender J, Danenberg KD, Metzger R, Schneider PM, Park J, Salonga D, Hölscher AH, Danenberg PV. Epidermal growth factor receptor and HER2-neu mRNA expression in non-small cell lung cancer Is correlated with survival. Clin Cancer Res. 2001. 7:1850–1855.
7. Baselga J. Targeting the epidermal growth factor receptor with tyrosine kinase inhibitors: small molecules, big hopes. J Clin Oncol. 2002. 20:2217–2219.

8. Thomas SM, Grandis JR. Pharmacokinetic and pharmacodynamic properties of EGFR inhibitors under clinical investigation. Cancer Treat Rev. 2004. 30:255–268.

9. Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004. 59:2 Suppl. 21–26.

10. Chang KH, Kim MS, Hong GW, Shin YN, Kim SH. Conversion of a murine monoclonal antibody A13 targeting epidermal growth factor receptor to a human monoclonal antibody by guided selection. Exp Mol Med. 2012. 44:52–59.

11. Kim SH, Park SY. Selection and characterization of human antibodies against hepatitis B virus surface antigen (HBsAg) by phage-display. Hybrid Hybridomics. 2002. 21:385–392.

12. Kim SH, Chun JH, Park SY. Characterization of monoclonal antibodies against carcinoembryonic antigen (CEA) and expression in E. coli. Hybridoma. 2001. 20:265–272.

13. Kim SH, Song SH, Kim YJ, Park SY. Expression and characterization of a recombinant Fab fragment derived from an anti-human alpha-fetoprotein monoclonal antibody. Mol Cells. 2001. 11:158–163.
14. Shin YW, Ryoo KH, Hong KW, Chang KH, Choi JS, So M, Kim PK, Park JY, Bong KT, Kim SH. Human monoclonal antibody against Hepatitis B virus surface antigen (HBsAg). Antiviral Res. 2007. 75:113–120.

15. Urlaub G, Mitchell PJ, Kas E, Chasin LA, Funanage VL, Myoda TT, Hamlin J. Effect of gamma rays at the dihydrofolate reductase locus: deletions and inversions. Somat Cell Mol Genet. 1986. 12:555–566.

16. Hong KW, Kim CG, Lee SH, Chang KH, Shin YW, Ryoo KH, Kim SH, Kim YS. A novel anti-EGFR monoclonal antibody inhibiting tumor cell growth by recognizing different epitopes from cetuximab. J Biotechnol. 2010. 145:84–91.

17. Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Res. 2002. 62:4132–4141.
18. Kim YS, Bhandari R, Cochran JR, Kuriyan J, Wittrup KD. Directed evolution of the epidermal growth factor receptor extracellular domain for expression in yeast. Proteins. 2006. 62:1026–1035.

19. Kim MS, Lee SH, Song MY, Yoo TH, Lee BK, Kim YS. Comparative analyses of complex formation and binding sites between human tumor necrosis factor-alpha and its three antagonists elucidate their different neutralizing mechanisms. J Mol Biol. 2007. 374:1374–1388.

20. Song MY, Park SK, Kim CS, Yoo TH, Kim B, Kim MS, Kim YS, Kwag WJ, Lee BK, Baek K. Characterization of a novel anti-human TNF-alpha murine monoclonal antibody with high binding affinity and neutralizing activity. Exp Mol Med. 2008. 40:35–42.

21. Cochran JR, Kim YS, Olsen MJ, Bhandari R, Wittrup KD. Domain-level antibody epitope mapping through yeast surface display of epidermal growth factor receptor fragments. J Immunol Methods. 2004. 287:147–158.

22. Burgess AW. EGFR family: structure physiology signalling and therapeutic targets. Growth Factors. 2008. 26:263–274.

23. Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987. 196:901–917.

24. Li S, Schmitz KR, Jeffrey PD, Wiltzius JJ, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005. 7:301–311.

25. Adams GP, Schier R, McCall AM, Simmons HH, Horak EM, Alpaugh RK, Marks JD, Weiner LM. High affinity restricts the localization and tumor penetration of single-chain fv antibody molecules. Cancer Res. 2001. 61:4750–4755.
26. Rudnick SI, Lou J, Shaller CC, Tang Y, Klein-Szanto AJ, Weiner LM, Marks JD, Adams GP. Influence of affinity and antigen internalization on the uptake and penetration of Anti-HER2 antibodies in solid tumors. Cancer Res. 2011. 71:2250–2259.

27. Kamat V, Donaldson JM, Kari C, Quadros MR, Lelkes PI, Chaiken I, Cocklin S, Williams JC, Papazoglou E, Rodeck U. Enhanced EGFR inhibition and distinct epitope recognition by EGFR antagonistic mAbs C225 and 425. Cancer Biol Ther. 2008. 7:726–733.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download