INTRODUCTION
The discovery of microRNAs (miRNAs), a new class of negative regulator that repress gene expression by pairing with their target messenger RNAs (mRNAs), has revealed a natural pathway for controling gene expression (1-3). There are hundreds of miRNAs encoded in the human genome and thousands of target mRNAs, which illustrates the important regulatory roles of miRNAs in cell development, differentiation. proliferation and apoptosis pathways (4,5). It is not surprising that deregulated miRNAs have been involved in the pathogenesis of many human disease (2,6-8). The recent development of technologies and compounds of identity and modulated miRNAs has opened new avenues for diagnosis, prognosis and therapeutic applications (6,9,10).
miRNAs are found in almost all species: virus, plants, nematodes, fly, fish, mouse, human, and are implicated in a wide array of cellular and developmental process (11). Extensive genome-wide expression profiling of cells and tissues in different stages of development or differentiation, metabolic conditions, and disease models using miRNA microarrays brought to the conclusion that unique miRNA profiles exist that are specific for the studied types of samples (1,11). These exciting but unexpected findings crystallized the hypothesis that genome-wide miRNA expression profiling could be used to profile tumors based on their origin and differentiation state, to help in diagnostic, prognosis, and for the use of miRNAs in therapeutic (11,12). Abnormal patterns of hematopoietic-specific miRNAs have been found in different types of cancer and miRNA-based gene therapy is being considered as a potential technology of choice in immunological disorders and cancer (11).
As mentioned above, miRNAs have recently emerged as important regulators of gene expression. They were formerly thought to mainly repress the translation of target mRNAs, but it has recently been shown that the main function of miRNAs in mammalian system is to decrease target mRNA levels (13). Recent evidence also suggests that the number of unique miRNA genes in human exceed 1,000, and may be as high as 20,000 and it is estimated that 20~30% of all human mRNA are miRNA targets (14). And so, it is probable that most mRNAs are controlled by miRNAs to some extent. The expression of miRNAs is highly regulated and they are therefore well placed to function as immunomodulators (15). More recently, miRNA are also proving to be an important link between the innate and adaptive immune systems, and their dysregulation might have a role in the pathogenesis of various diseases (1,2,15).
Importantly, it has been increasingly reported that miRNAs are associated with disease. However, the pattern among the miRNA-disease association remains largely unclear. In order to dissect the patterns of miRNA-disease associations, Lu et al. performed a comprehensive analysis to the human miRNA disease association data, which is manually collected from publication (16). They built a human miRNA association disease network. Interestingly, miRNAs tend to show similar or different dysfunctional evidence for the similar or different disease clusters, respectively (16). In addition, they also found that there is a negative correlation between the tissue-specificity of a miRNA and the number of disease it associated and that there is an association between miRNA conservation and disease (16). Furthermore, they uncovered that miRNAs associated with the same disease tend to emerge as predefined miRNA groups. These findings can not only provide help in understanding the association between miRNAs and human diseases, but also suggest a new way to identify novel disease-associated miRNAs (16).
Their biological importance, initially demonstrated in cancer, was also more recently discovered in many other pathologies like Alzheimer's disease (AD), Parkinson's disease (PD), viral infections, diabetes, and myopathies (17). They summarized miRNA profiling studies in human diseases and discuss the newly discovered link between miRNAs and drug response. An understanding of how miRNAs influence the body's response to certain drugs and how these affect the expression of miRNAs, will be of key importance in developing drugs with greater safety and efficacy (17).
miRNA expression can be induced or expressed by a variety of stimuli and mechanisms. These stimuli include direct transcriptional activation or repression from transcriptional enhancers, epigenetic modifications of the genome, genomic amplification or deletion, cellular stress and inflammatory stimuli (1,13,18). The effect of inducing or repressing miRNA expression can influence most biological processes, including cell fate specification, cell proliferation, DNA repair, DNA methylation and apoptosis and provide pro-inflammatory or anti-inflammatory stimuli (8,19). The biological effect of a specific miRNA will depend on the cellular environment in which it is expressed its turnover rate and the target sequence that the miRNA can bind (1,13). Since a single miRNA can bind its target sequence with imperfect complementarity, a specific miRNA can have many potential targets (1,13). This allows a single type of miRNA to simultaneously regulate the translation of many genes in multiple pathways. In this manner, relatively small changes in miRNA expression can lead to modest changes in the levels of multiple proteins and collectively these can add up to large changes in biology (20-22). Selbach et al. demonstrated that, in addition to down-regulating mRNA levels, miRNAs also directly repress translation of hundreds of genes and that a miRNA can, by direct or indirect effects, tune protein synthesis from thousands of genes (20). miRNAs also have an essential role in both the innate and adaptive immune system. Proper miRNA expression is required for correct differentiation of immune cells (23). Immune responses are symphonies of molecular and cellular interactions, with each player doing its part to produce the composite behavior we see as effective host defense, or when discoordinated, as immunopatholgy or immunodeficiency (22). It is therefore not surprising that they have been implicated in various human diseases like cancer (2,19,24-28). cardiovascular diseases (5,6,29,30), neurological diseases (7,31), psychiatric diseases (10,32,33), autoimmune diseases (9,34-38), skin diseases (39,40-42), asthma (43-45) and microbial infection (1,46-50). In this review, I briefly summarize the current knowledge of roles of miRNA in various human cancers and cardiovascular disease.
miRNA BIOGENESIS
All miRNAs are processed and matured through a complex biogenesis process involving multiple protein catalysis accessory proteins, and macromolecular complexes following a coordinated series of event. The reader is referred to excellent reviews for detailed discussion of miRNA biogenesis and its regulation (2,51-54). miRNAs constitute a class of small endogenous noncoding RNAs of 19-23 nucleotides that negatively regulate gene expression (51,52). They are an abundant class of gene regulatory molecules in multicellular organisms and modulate the expression of many protein-coding genes (51,53,54). They are transcribed as a huge double-stranded primary transcript (pri-miR) by RNA polymerase II. Subsequently, nuclear enzyme Drosha (also known as ribonuclease III) and Pasha convert this precursor into a double-stranded miRNA precursor of ~70 nulcleotide (pre-miR), which is next transported into the cytoplasm by a mechanisms involving the protein Exportin 5 (2,15,51,55). Finally, Dicer enzyme processes this precursor into the 22-nucleotide double-stranded miRNA. This duplex is then unwinded, and the leading strand ("guide strand"), one of the two strands, is incorporated into the RNA-induced silencing complex (RISC), which is comprise Agonaute and other proteins (2,55,56). miRNAs incorporated in the RISC are able to bind to the 3' untranslated region (UTR) of target mRNAs causing a block of translation or mRNA degradation depending on the level of complementarity (51,53). The other strand so-called "passenger strand" is degraded (2,40,51). Recent studies have clearly demonstrated that miRNAs play critical roles in several biologic processes, including differentiation, development, cell growth, and apoptosis, by regulating gene expression through either the inhibition of mRNA translation or the induction of its degradation (15,18,19). The analysis of human neoplasias of different tissue origins has shown deregulated miRNA expression (57,58).
miRNA FUNCTIONS
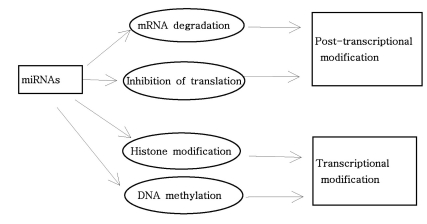
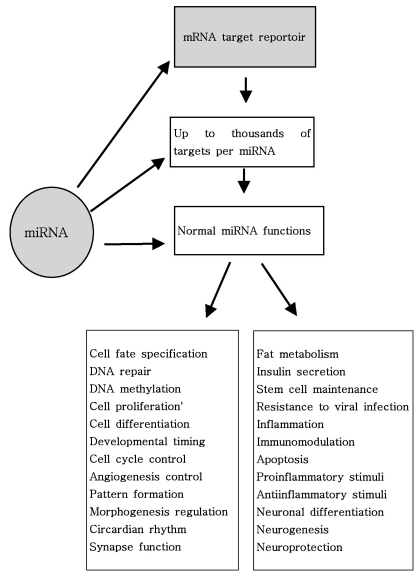
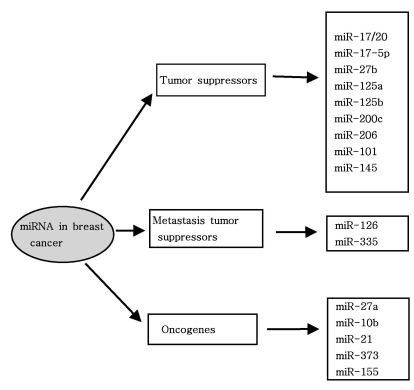
As mentioned above, miRNAs recognize their target mRNAs based on sequence complementarity and act on them to cause the inhibition of protein translation by degradation of mRNA (51,59). It becomes evident that miRNA must be involved in developmental timing, cell death, cell proliferation, hematopoiesis, patterning of nervous system and other normal cellular homeostasis (51,60,61). As shown in Fig. 1, it is becoming clear that miRNAs is only regulated gene expression at the post-transcriptional level, but they are also capable of modifying chromatin (60,62,63). As shown in Fig. 2, participation of miRNAs in essential biological processes has been consistently proven (60,62-66), such as cell proliferation control (miR-125b and let-7), hematopoietic B-cell lineage fate (miR-181), B-cell survival (miR-15a and miR-16-1), brain patterning (miR-430), pancreatic cell insulin secretion (miR-375), and adipocyte development (miR-143). Furthermore, with the development of new techniques for genome-wide screening of miRNA expression, abnormal levels of miRNA were identified in various diseases with respect with normal counterpart (58,59). Given the importance of miRNAs in regulating cellular differentiation and proliferation, it is not surprising that their misregulation is linked to cancer. In cancer, as shown in Fig. 3, miRNAs function as regulatory molecules, acting as oncogenes or tumor suppressors (67-71). It is interesting to note that some miRNAs may have dual functions as both tumor suppressor and oncogenes (72). Amplification or over-expression of miRNAs can down-regulate tumor suppressors or other genes involved in cell differentiation, thereby contributing to tumor formation by stimulating proliferation, angiogenesis, and invasion, i.e., they act as oncogenes (59,65,66). Similarly, miRNAs can down-regulate different proteins with oncogenic activity, i.e., they act as tumor suppressor (59,65,66). Because of the small size of miRNAs, loss-of-function or gain-of-function point mutations represent rare event (58). miRNA activity can be influenced by the reposition of other genes closed to promotors/regulatory regions of miRNAs (as is the case of miR-142s-cMYC translocation), or by the relocalization of an miRNA near other regulatory elements (64). The overall effects in the case of miRNA inactivation is the over-expression of target mRNAs, while miRNA activation will lead to down-regulation of target mRNAs involved in apopotosis, cell cycle, invasion, or angiogenesis (63,64).
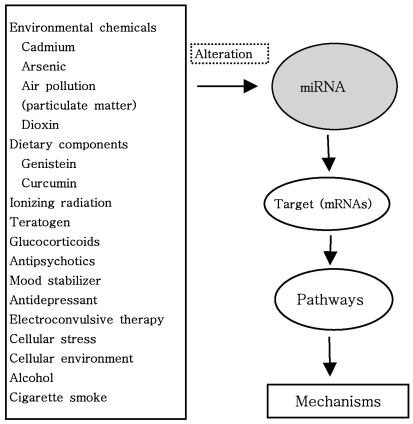
Very importantly, as shown in Fig. 4, miRNAs are altered or induced by both environmentally regulated early life developmental factors through epigenetic and miRNA mechanisms, and genetic polymorphisms including protein or miRNA genes (73). A growing body of experimental evidence suggests that miRNAs are altered by environmental chemicals such as cadmium, arsenic, dioxin, nickel, chronium, methylmercury, benzene and air pollution (74). Surprisingly, exposure to air pollution, particularly to particulate matter, has been associated with increased morbidity and mortality from cardiorespiratory disease, as well as with lung cancer risk (74). Additionally, it is interesting that a growing body of literature suggest that miRNAS can be altered by environmental and dietary factors such as genistein, curcumin (diferuloylmethane, naturally occurring flavinoid and proapoptotic compound derived from the rhizome of Curcuma longa), retinoic acid, folate as well as teratogen and ionizing radiation (74-81). It has been known that miRNAs are altered by vinyl carbamate (VC) and the cancer preventive effect of cruciferous vegetables is attributed to their different phytochemical constituents (82). One of the most important anti-carcinogenic phytochemicals contained in these vegetables is indole-3-carbinol (I3C), an enzymatic breakdown product of indole glucosiniolates (82). Melkamu et al. examined if the chemopreventive agent I3C reversed VC-induced deregulation of miRNA levels in lung tissues of female A/J mice (82). They reported that miRNAs are deregulated during VC-induced mouse lung tumorigenesis and their levels are modulated by I3C (82). A recent another study demonstrated that teratogen-induced limb dysmorphogenesis may be associated with alteration in miR-34 and miR-125b expression and that p-53-independent mechanisms exist contributing to teratogen-induced activation of miR-34a and miR-34c: some miRNAs act to protect embryos, whereas other miRNA boost a teratogen-induced process of maldevelopment to induce embryonic death (79). Interestingly, some miRNAs, which have been shown to change expression in response to cellular stimulation, act as negative or positive feedback regulators and they also may have key roles in mediating cross talk between inflammatory mediators (22).
Interestingly, miRNAs are also regulated by stress and glucocorticoids and during the cellular stress response (73). miRNAs have the capacity to change from translation suppressor to activator by forming new interactions between miRNA/Argonaute complexes and RNA-binding protein that alter their subcellular localization (73). Because of this modified activity, miRNAs could provide a pivotal role in mediating cellular adaptation to stress (73). In particular, miRNAs have been implicated in the way cells respond to oxidated stress, nutrient deprivation, and DNA damage (73). Chronic psychosocial stress is known to have adverse physiological effects that contribute to cardiovascular disease, impaired immune function, inflammatory disease, and neuronal function (73). Glucocorticoids are one of the prominent mediators of cellular stress effects on neural function and behavior, and are known to structurally alter brain cytoarchitecture in regions that contribute to cognition, memory and emotion (73).
As shown in Fig. 2, Nelson et al. summarized recent studies that have revealed intriguing novel aspects of miRNA function (83): 1) some miRNA are edited post-transcriptionally, and thus their targets can be profoundly altered, 2) some human miRNAs are targeted to the nucleus for as yet unknown reasons, 3) miRNA are shown to alter mRNA splicing and/or DNA methylation in trans, 4) miRNAs (like siRNAs) may recognize mRNAs outside of the 3'UTR, opening up new possibilities for complex gene expression regulation, 5) viral-derived miRNAs/siRNAs can play important roles in host cell biochemistry, 6) under some circumstances, miRNAs can increase translation of targeted mRNAs, and 7) numerous cell- and organism-level functions as diverse as immunomodulation, endocrine function, circadian rhythms, metabolic pathways, limb morphogenesis, and angiogenesis have been hypothesized to be regulated by miRNAs. In addition, these alteration can be caused by various mechanisms, including deletion, amplifications or mutation involving miRNA loci, epigenetic silencing or dysregulation of transcription factors that target specific mRNA (25,58). As mentioned above and as will be discussed further, recently, a growing body of experimental evidence strongly suggests an unexpected mechanism of miRNA involvement in various human diseases ranging from cancer to psychiatric diseases (25,28).
CANCER
There has been an explosion of literature focusing on the role of regulatory T (Treg) cells in cancer immunity and miRNAs are highly expressed in Treg cells (15,84-87). And, over the past several years it has become clear that alteration in the expression of miRNA genes contribute to the pathogenesis of most-if not all- human malignancies (25,58). These alteration can be caused by various mechanisms, including deletions, amplifications or mutations involving miRNA loci, epigenetic silencing or the dysregulation of transcription factors that targets specific miRNAs (25,58). Because malignant cells show dependence on the dysregulated expression of miRNA genes, which in turn control or are controlled by the dysregulation of multiple protein-coding oncogenes or tumor suppressor genes, these small RNAs provide important opportunities for the development of future miRMA-based therapies (24,88).
Within the past few years, studies on miRNA and cancer have burst onto the scene. Profiling of the miRNome (global miRNA expression levels) has become prevalent, and abundant miRNome data are currently available for various cancers (15,57,89). As mentioned, miRNA are a recently discovered class of small non-coding RNAs that regulate gene expression (2,24,90). Mature miRNA are the results of sequential processing of pri-miRNAs mediated by two RNAse III enzymes, Drosha and Dicer (15,51-55). Mature miRNAs negatively regulate protein expression of specific mRNAs by either translational inhibition or mRNAs degradation (2,4,58). miRNA can contribute to cancer development and progression and are differentially expressed in human cancers (15,25,89). Calin and Croce. also reported that miRNA-expression profiling of human tumors has identified signatures associated with diagnosis, staging, progression, prognosis and response to treatment (58). From a large-scale miRnome analysis on 540 samples including lung, breast, stomach, prostate, colon and pancreatic tumors, Volinia et al. identified a solid cancer miRNA signature composed by a large portion of over-expressed miRNAs (57). They reported that among these miRNAs are some with well characterized cancer association, such as miR-17-5p, miR-20a, miR-21, miR-92, miR-106, and miR-155 (57). The predict targets for the differentially expressed miRNAs are significantly enriched for protein-coding tumor suppressor and oncogene. A number of the predicted targets, including the tumor suppressor RB1 (retinoblastoma 1) and TGFBR (transforming growth factor, beta receptor II) genes were confirmed experimentally (57). Their studies indicate that miRNAs are extensively involved in cancer pathogenesis of solid tumors and support their function as either dominant or recessive cancer genes (57).
Importantly, miRNA deficiencies or excess have been correlated with a number of clinically important diseases ranging from myocardial infarction to cancers (67). The loss or gain of miRNA function can be caused by a single point mutation in either the miRNA or its target or by epigenetic silencing of pri-miRNA transcription units (67). Interestingly, a high-throughput analysis of miRNA expression in cancer demonstrated that some miRNA are over-expressed in cancer, while others are markedly reduced in malignant tissue (67). These correlative data suggest that miRNAs function as both oncogenes and tumor suppressors (67-71). Numerous studies in cancer cell lines show a direct functional link between aberrant miRNA expression and particular tumor types (58,67,69,91). As aforementioned, recent studies also show that some miRNAs regulate cell proliferation and apoptosis processes that are important in cancer formation (69). By using multiple molecular technique, which include Northern blot analysis, real-time PCR, miRNA microarray, up- or down-expression of specific miRNAs, it was found that several miRNAs were directly involved in human cancers, including lung, breast, brain, stomach, liver, colon, prostate, thyroid, pancreas, ovary cancer and leukemia (4,19,67,69,92,93). In addition, some miRNAs may function as oncogene or tumor suppressor (67,69-71). More than 50% of miRNA genes are located in cancer-associated genomic regions or in fragile sites, suggesting that miRNAs may play a more important role in the pathogenesis of a limited range of human cancers than previously thought (67,71). Interestingly, over-expressed miRNA in cancers, such as miR-17-92, may function as oncogene and promote cancer development by negatively regulating tumor suppressor genes and/or genes that control cell differentiation or apoptosis (69,91). Under-expressed miRNAs in cancers, such as let-7, function as tumor suppressor genes and may inhibit cancer by regulating oncogenes and/or genes that control cell differentiation or apoptosis (69,70,91). miRNA expression profiles may become useful biomarkers for cancer diagnosis and miRNA therapy could be a powerful tool for cancer prevention and therapeutics in not distant future (69,70,94,95). Moreover, since misregulation of miRNA has been associated with various cancers, the identification of specific regulators of miRNAS will be helpful in developing new therapeutic agents.
BREAST CANCER
Interestingly, miRNA aberrant expression has been found in human breast cancer, where miRNA signature were associated with specific clinicobiological features (93). Compared with normal breast tissue, miRNAs are aberrantly expressed in human breast cancer. Overall miRNA expression could clearly separate normal versus cancer tissues, with the most significantly deregulated miRNAs being miR-125b, miR-145, miR-21, and miR-155 (93). Interestingly, Tavazoie et al. showed that restoring the expression of some miRNAs in malignant cells suppresses lung and bone metastasis by human cancer cells in vivo (96). Of these miRNAs, miR-126 restoration reduces overall tumor growth and proliferation, whereas miR-335 inhibits metastatic cell invasion (96). They also reported that miR-335 regulates a set of genes whose collective expression in a large cohort of human tumors is associated with risk of distal metastasis and that miR-335 suppresses metastasis and migration through targeting of the progenitor cell transcription factor SOX4 and extracellular matrix component tenascin C (96). Moreover, they demonstrated that expression of miR-126 and miR-335 is lost in the majority of primary breast tumors from patients who relapse, and the loss of expression of either miRNA is associated with poor distal metastasis-free survival. Thus, they noted that miR-335 and miR-126 are identified as metastasis suppressor miRNAs in human breast cancer (96).
Recently, miRNAs are thought to regulate invasion via direct interaction with target genes within cells (26). A study showed that miR-17/20 cluster inhibit cellular migration and invasion of nearby cells via heterotypic secreted signals in breast cancer, indicating that the findings not only reveal an anti-invasive function of miR-17/20 cluster in breast cancer, but also identify a heterotypic secreted signal that mediates the miRNA regulation of tumor metastasis (26).
As discussed, the presence of Treg cells in breast cancer marks an invasive phenotype and poor prognosis (97). In addition to their immunosuppressive role in antitumoral responses, CD4+Treg cells contribute to mammary tumor metastasis through the expression of receptor activator of nuclear factor-κB ligand (RANKL) and its receptor RANK (97). Tan et al. currently examined whether RANKL and RANK are involved in mammary/breast cancer metastasis (97). They found that tumor-infiltrating Treg cells stimulate mammary cancer metastasis through RANKL-RANK signalling and that CD4 Treg cells are the main products of RANKL in breast cancer tumors (97). They also reported that most RANK-producing cells expressed Foxp3 and that their results are consistent with the adverse impact of tumor-infiltrating CD4+ or Foxp3+ T cells on human breast cancer prognosis. These results suggest that the targeting of RANKL-RANK can be used in conjunction with the therapeutic elimination of primary breast tumors to prevent recurrent metastastic disease (97).
PANCREATIC CANCER
Pancreatic cancer is the leading cause of cancer-related death and the prognosis for pancreatic cancer is the worst of all cancers with high mortality, a mortality/incidence ratio of 0.99 (92). The incidence of pancreatic cancer in the United States is ~9 per 100,000. These discouraging numbers, reflecting the increasing rates of incidence and death, are due to the lack of improvement in detection and diagnosis strategies and the paucity of breakthroughs in treatment regimens (92). A miRNA expression signature has been identified that is associated with pancreatic cancer and this has been accomplished with the application of real-time PCR profiling of 200 miRNA precursors on specimens of human pancreatic adenocarcinoma, paired benign issue, normal pancreas, pancreatitis and cell lines (92). Lee et al. (92) showed that one hundred miRNA precursors were aberrantly expressed in pancreatic cancer or desmoplasia, including miRNAs previously reported as differentially expressed in other human cancers (miR-155, miR-21, miR-221, and miR-222) as well as those not previously reported in cancer (miR-376a and miR-301). They also demonstrated that most of the top aberrantly expressed miRNAs displayed increased expression in tumor and that three of the top differentially expressed miRNAs (miR-221, miR-221, miR-301) were localized to tumor cells and not to stroma or normal acini or ducts (92). They noted that aberrant miRNA expression may offer new clues to pancreatic tumorigenesis and may provide diagnostic biomarkers for pancreatic adenocarcinoma (92). It is well established that many tumor suppressor genes in human cancer are silenced by chromatin alterations, including promoter methylation and histone deacetylation (98). Lee at al. treated two human pancreatic cancer cell lines (MiaPACA-2 and PANC-1) with the demethylating agent, 5-aza-2'-deoxycytidine or histone deacetylase inhibitor, trichostatin A, as well as the combination of the two (98). They assessed expression of miRNAs in control and treated cell lines using a custom microarray platform (98). They found that fourteen miRNAs were up-regulated two-fold or greater in each of the cell lines following exposure to both chromatin-modifying agent, including 5 miRNAs that were in common (miR-107, miR-103, miR-29a, miR-29b and miR-320) to both MiaPACA-2 and PANC-1 (98). Enforced expression of miR-107 in the cell lines was down-regulated in vitro growth, and this was associated with repression of the putative miR-107 target, cyclin-dependent kinase 6, thereby providing a functional basis for the epigenetic inactivation of this miRNA in pancreatic cancer (98). Additionally, pancreatic ductal adenocarcinoma (PDAC) is known for its very poor overall prognosis (99). Therefore, accurate early diagnosis and new therapeutic modalities are urgently needed. Recently, using 377 feature miRNA array, Szafranska et al. investigated miRNA expression in normal pancreas, chronic pancreatitis, and PDAC as well as PDAC-derived cell lines (99). They found that the expression of miR-216 and miR-217 and lack of expression of miR-133a were identified as characteristic of pancreas tissue. The authors also identified 26 miRNAs most prominently misregulated in PDAC. Their data provide novel insights into the miRNA-driven pathophysiological mechanisms involved in PDAC and offers new candidate targets to be exploited both for diagnosis and therapeutic strategies (98).
A recent study showed that pancreatic cancer tissues or cell lines have a unique miRNA profiling pattern at the individual basis as compared with relatively normal pancreatic tissues or cells as well as pancreatitis tissue (100). This study also showed that eight miRNAs were significantly up-regulated in most pancreatic cancer tissue and cell lines, including miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b, and miR-95. Interestingly, the incidence of up-regulation of these eight genes between normal controls and tumor cells or tissues was ranging from 70% to 100% (100). Interestingly, miR-21 is relatively over-expressed in glioblastoma multiforme, cervical cancer, beast cancer, and many other solid tumor types (55). Increased expression of miR-21 appears to contribute to decreased apoptosis in malignant cell (55). Similarly, the locus containing seven miRNAs of the miR-17-92 polycistron cluster at human chromosome 13q31 is amplified in a variety of B cell lymphomas, nasal NK/T cell lymphoma, and solid tumor (55). Conversely, some miRNAs can act as tumor suppressor, genes whose deletion or mutation helps cell along the multi-step process of tumorigenesis (55). Circumstantial evidence for this possibility first emerged in early miRNA profiling experiments where tumor samples appeared to have lower overall levels of miRNA expression than normal tissue sample, both in human clinical material and tissue from murine cancer model (55).
In addition, tumor phenotype associated with extensive disease, such as invasion of surrounding tissue and metastasis, are also under the influence of miRNA controls (55). As mentioned, experimental evidence has shown that miRNAs can play roles as oncogenes or tumor suppressor genes, suggesting their contribution to cancer development and progression (69,70,91). Expression profile of human miRNA demonstrated that many miRNAs are deregulated in cancers and are differentially expressed in normal tissues and cancers (69,70,91). Therefore, miRNAs profiling is used to create signature for a variety of cancer, indicating that the profile will help further establish molecular diagnosis, prognosis and therapy using miRNAs (71).
COLORECTAL CANCER (CRC)
Changes in the expression profiles of miRNAs have been observed in a variety of human tumors, including CRC (4). Several investigators have also described the ability of miRNA expression profiles to predict prognosis and response to selected treatment in CRC patients, and support diagnosis of CRC among cancer of unknown primary site (4). miRNA's occurrence has been repeatedly observed also in serum and plasma, and miRNAs as novel minimally invasive biomarkers have indicated reasonable sensitivity for CRC detection (4). Two approach are applied today to investigate the connection between miRNAs and CRC (4).: functional and profiling studies. On one hand, miRNAs seem to regulate many known oncogenic and tumor suppressor pathways involved in the pathogenesis of CRC. Many protein involved in key signaling pathway of CRC, such as members of Wnt/β-catenin and phosphatidylinositol-3-kinase (PI-3-K) pathways, KRAS, p53, extracellular matrix regulators as well as epithelial-mesenchymal transition (EMT) transcription factors, are altered and seem to be affected by miRNA regulation in CRC (4).
Recent studies have shown that, in cancer, expression of some miRNAs cells is silence in association with CpG island hypermethylation (101). To identify epigenetically silenced miRNAs in CRC, Toyota et al. screened for miRNAs induced in CRC cells. They found that miR-34b and miR-34c, two components of the p53 network, are epigenetically silenced in CRC (101). They also found that the miR-34b/c CpG island is a bidirectional promoter which drives expression of both miR-34b/c and B-cell translocation gene 4 (BTG4). These results suggest that miR-34b/c and BTG4 are novel tumor suppressors in CRC and that the miR-34b/c CpG island is a frequent target of epigenetic silencing in CRC (101).
Recently, Ng et al. investigated whether plasma miRNA could discriminate between patient with and without CRC and found that of the panel of 95 miRNAs analysed, five were up-regulated both in plasma and tissue sample and that all the five miRNAs were validated on the plasma of 25 patients with CRC and 20 healthy controls (102). They also found that both miR-173p and miR-92 were significantly elevated in the patients with CRC and that the plasma levels of these markers were significantly reduced after surgery in 10 patients with CRC (102). Further validation with an independent set of plasma sample indicated that miR-92 differentiates CRC from gastric cancer, inflammatory bowel disease (IBD) and normal subjects, indicating that miR-92 can be a potential non-invasive molecular marker for CRC screening (102). Recently, Wang et al. investigated the miR-31, miR-143 and miR-145 expression in 98 primary CRC specimens, along with the corresponding normal mucosa specimens, and analyzed the relationship of their expression with clinicopathological features (103). Their results showed the miR-31 expression was up-regulated in CRC compared to normal mucosa. They also showed that the miR-31 over-expression may be involved in the development and progression of CRC and that the miR-143 and miR-145 may play a certain role in the development of colon and/or rectal cancers but not in progression of the disease (103). Recent reports have highlighted the oncogenic aspects of miR-125b. However, the clinical significance of miR-125b in gastrointestinal cancers has not been sufficiently investigated. Currently, however, Nishida et al. analyzed miR-125b expression in CRC cases (104). Evaluating miR-125b expression in 89 CRC cases, the authors found that miR-125b is directly involved in cancer progression and is associated with poor prognosis in human CRC (104). Their findings suggest that miR-125b could be an important prognostic indicator for CRC patients. Huang et al. measured the levels of 12 miRNAs in plasma samples from patients with advanced colorectal neoplasia (carcinomas and advanced adenomas) and healthy controls using real-time RT-PCR (105). They found that plasma miR-29a and miR-92a have significant diagnostic value for advanced neoplasia. More importantly, these 2 miRNAs also could discriminate advanced adenomas from controls (105). These data suggest that plasma miR-29a and miR-92a have strong potential as novel noninvasive biomarkers for early detection of CRC.
Importantly, by modulating oncogenic and tumor suppressor pathways miRNAs could, in principle, contributes to tumorigenesis (90). And recurrent genetic and epigenetic alterations of individual miRNAs are found in some tumors (90). miRNAs that are amplified or over-expressed in cancer could act as oncogenes, and a number of putative oncogenic miRNAs have been proposed (2,55,58,67). An interesting case is represented by miR-155, which is up-regulated in several hematopoietic malignancies and tumors of the breast, lung, and pancreas (2,58,67,90). Another notable member of the family of oncogenic miRNA is the miR-17-92 cluster (58,89,90).
The role of miRNAs in tumorigenesis underscores their value as mechanism-based therapeutic targets in cancer (95). Similarly, unique patterns of altered levels of miRNA production provided fingerprints that may serve as molecular biomarkers for tumor diagnosis, classification, prognosis of disease-specific outcomes and prediction of therapeutic response (94,95). Recent studies demonstrated that the best characterized tumor-suppressor miRNAs are miR-15a and miR-16 and that B cell chronic lymphocytic leukemia (CLL) is the most common adult leukemia in developed countries and is universally associated with the loss of chromosomal region 13q14 (58). The best characterized oncogenic miRNAs are the miR-17 cluster which comprises a group of six miRNAs (miR-17-5p, miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92) at 13q31-32, a chromosomal region amplified in large B-cell lymphoma, follicular lymphoma, mantle cell lymphoma and primary cutaneous B-cell lymphoma (95).
The "classic" view of molecular oncology indicates that cancer is a genetic disease involving tumor suppressor and oncogenic proteins (106). However, recent years, it has been demonstrated that miRNA are involved in human tumorigenesis, thus revealing a new layer in the molecular architecture of human cancer (106). Gene expression studies revealed that hundreds of miRNAs are deregulated in cancer cells and functional studies clarified that miRNAs are involved in all the molecular and biological processes that drive tumorigenesis (106).
LUNG CANCER
Lung cancer is the leading cause of death from cancer in the world (107). miRNA have multiple functions in lung development, and abnormal expression of miRNAs could lead to lung tumorigenesis (107). The different expression profiles of miRNAs in lung cancer, and the stability of miRNAs in serum, all together make them as new potential clinical biomarkers for diagnosis and prognosis (107). As mentioned, moreover, miRNAs may serve as either novel potential targets acting directly as oncogenes (e.g. miR-17-92 cluster) or directly therapeutic molecules working as tumor suppressor genes (e.g. let-7 family) (107). Twenty-three of the 30 most highly expressed miRNAs shared in adult lung from mice and human. The miR-26, let-7, miR-29, miR-30 and miR-99 were expressed highly in both species (107). Importantly, Wang et al. reported 123 miRNAs that may have potential roles in lung cancer (107). In the same paper, they reviewed dysregulation of various types of miRNA in development of lung and lung cancer (107).
In order to characterize the function of hsa-miR-125a-3p/5p in invasion and metastasis of non-small cell lung cancer (NSCLC), Jiang et al. investigated the relationship between hsa-miR-125a-3p/5p expression and lymph node metastasis in NSCLC tissue (108). They found that hsa-miR-125a-3p and hsa-miR-125a-5p play distinct roles in regulation of invasive and metastatic capabilities of lung cancer cells and lymph node metastasis in NSCLC, indicating that miR-125a family members play a important role in the development of NSCLC (108). As discussed, miRNA have been recently implicated in several carcinogenic processes, where they can act either as oncogenes or as tumor suppressors (67,68). This is the case in lung cancer, i.e. the leading cause of cancer death in Western countries, in which about 40-45 miRNAs have been found to be aberrantly expressed, thereby constituting a specific miRNA signature (89). Some transcript of the let-7 family that are significantly down-regulated in lung tumors have been identified as tumor suppressors through their ability to control several oncogenic pathway, including the RAS pathway (89).
The pattern of miRNA expression can be correlated with cancer type, stage, and other clinical variables, so miRNA profiling can be used as a tool for cancer diagnosis and prognosis (109). miRNAs play roles in almost all aspects of cancer biology, such as proliferation, apoptosis, invasion/metastasis, and angiogenesis. It is expected that more miRNAs will emerge as players in the etiology and progression of cancer (109).
GASTRIC CANCER
Recent studies have shown that some miRNAs play roles as tumor suppressor or oncogene in gastrointestinal cancer and that miRNA expression is regulated by different mechanisms including transcription factor binding, epigenetic alteration, and chromosomal abnormalities (18,19,90). miRNA expression profiling may be powerful clinical tool for cancer diagnosis and regulation of miRNA expression could be a novel strategy for chemoprevention of human gastrointestinal cancer (110). To study the role of miRNAs in gastric cancer, Yao et al. analyzed the expression profile of 847 miRNAs in Chinese patients with gastric cancer (111). The results from the miRNA microarray analysis were validated by real-time RT-PCR. They found that a total of 24 miRNAs with a more than 2-fold change were differentially expressed between normal gastric tissue and gastric cancer (111). Of these, 22 miRNAs, including miR-223, miR-106b, miR-147, miR-34a and others, were significantly up-regulated in gastric cancer, whereas only miR-638 and miR-378 were significantly down-regulated in gastric cancer compared to normal gastric tissue (111). These results show that miRNAs are deregulated in gastric cancer, suggesting the involvement of these genes in the development and progression of gastric cancer.
Interestingly, a recent study showed that miR-21 is consistently up-regulated in solid human cancers, including the stomach, as compared with matching noncancerous tissue (48). Direct targets of miR-21 have been identified, with all of them being tumor suppressors. miR-21 was over-expressed in gastric cancer tissue sample and cell lines, as well as in chronically Helicobacter pylori-infected gastric epithelium tissue, as opposed to noninfected tissue (48).
With the capacity of miRNA to alter the survival and death of T and B cells, control over miRNA expression is essential to prevent adaptive immune cells from unregulated proliferation (24). miRNA can act both as 'oncomirs' and tumor suppressors, and thus dysregulation of miRNA in lymphocytes can cause malignancies (24,67,68). With strong evidence of miRNA-mediated control over T cell and B cell development and well-established roles in the regulation of genes controlling apoptosis and the cell cycle, it is not surprising that there is growing evidence for a causative role of miRNA in the development of malignancies of the adaptive immune system (24). Analysis of miRNA expression profiles has demonstrated altered expression in numerous hematologic malignancies (24). The miRNA signature identified from these clinical studies have potential value as prognostic parameters for cancer progression. Futhermore, functional characterization of these miRNA may provide insights into the potential of manipulation selective miRNAs as a novel therapeutic means to treatment of hematologic cancer (24,112). The non-random genomic distribution of miRNA, with tenfold enrichment in fragile sites, suggests that at least some of these expression changes will be causative in tumor development (113). To date, many miRNAs with oncogenic activity in hematologic malignancies have been reported. These so-called 'oncomirs' such as the miR-17~92 cluster, miR-21 and miR-155 are often found over-expressed in malignant tissues. For example, the mir-17~92 cluster is a target of genomic amplificaion of 13q31 that occurs in Burkitt's lymphoma, diffuse large B-cell lymphoma (DLBCL), mantle cell lymphoma and follicular lymphoma (114). In addition to the miR-17~92 cluster, miR-155 is one of the oncomirs which has been well characterized in the adaptive immune system in both healthy development and malignancy, including B cell lymphoma, Hodgkin's lymphoma, DLBCL and Burkitt's lymphoma (115). In contrast to the oncomirs described above, other miRNA function as potent tumor suppressors. The loss of such miRNAs are frequently associated with T cell and B cell lymphoma and leukemia (115). One example is miR-15a and miR-161, two miRNA clustered in the 13q14 region. In more than 50% of CLL patients, this region was found deleted (116). Similarly, miR-29 and miR-181 are down-regulated in chronic lymphocytic leukemia (CLL) and have both been shown to function as tumor suppressor. These miRNAs normally suppress proliferation by targeting pro-survival and pro-proliferation genes including BCL-2, TCL-1, MCL-1 and CDK6 (117).
Recent studies have shown that miRNAs have unique expression profiles in cells of the innate and adaptive immune systems and have pivotal roles in the regulation of both cell development and functions (2,15). Furthermore, when miRNAs are aberrantly expressed they can contribute to pathological conditions involving the immune system, such as cancer and autoimmunity (2,15). As discussed, miRNAs directly modulate the concentration of many regulatory proteins that are required for normal development and function of the immune system. These proteins have been linked to immunological disease, in which the miRNAs are found to be mutated or their expression levels dysregulated, consequently triggering altered or impaired function (15,113). miRNA levels are also dysregulated in diseases of immunological origins and many are known to be encoded near fragile sites in the genome (113). They have also been shown to be useful as diagnostic and prognostic indicators of disease type and severity.
Acute inflammatory response is usually beneficial, especially in response to microbial infection and tissue damage (118). A well-regulated inflammatory response can also be anti-tumorigenic and have a role in tumor suppression (118). Chronic inflammation and infection, however, is detrimental and, among other deleterious effects, will frequently predispose cells for an oncogenic transformation (25). Key mediator of inflammation-induced cancer include nuclear factor kappa B (NF-κB), reactive oxygen and nitrogen species, inflammatory cytokines, prostaglandins and specific miRNAs (25). The potential to therapeutically regulate miRNA levels may offer new avenues for cancer treatment and possibly in regulating the immune system. Inhibiting oncogenic miRNA or reintroduction of tumor suppressor miRNAs may serve as useful strategies to treat cancer (4,18). Reintroduction of tumor suppressor miRNAs can cause apoptosis or senescence in malignant cell and provide new avenues for developing cancer therapeutics (4,18). A recent study clearly demonstrated this potential in mouse models of hepatocelluar carcinoma (HCC) patients (119). It has been shown that mi-26a is a tumor suppressor miRNA that is reduced in HCC and that decreased levels of miR-26a have been associated with poor prognosis and predictive to the therapeutic response to IFNα in HCC patients (25,119). Within the next several years, we will know if miRNA-based therapeutics, alone or in combination with other modalities, will be clinically useful treatment for various cancers and immune system disorders.
THYROID CANCER
A recent study investigated the expression patterns of miRNA in all major types of thyroid tumors, including tumors carrying distinct oncogenic mutations, and the utility of miRNA profiling for the preoperative diagnosis of thyroid nodules (120). It was reported that a set of seven miRNAs (miR-187, miR-221, miR-222, miR-146b, miR-155, miR-224, and miR-197) were most differentially over-expressed in thyroid tumors (120). Moreover, it was demonstrated that various histopathological types of thyroid tumors have distinct miRNA profiles, which further differ within the same tumor type, reflecting specific oncogenic mutations (120). These results may suggest that a limited set of miRNAs can be used diagnostically with high accuracy to detect thyroid cancer in the surgical and preoperative samples (120).
The thyroid gland is composed of two distinct hormone-producing cell types: follicular cells and parafollicular C-cells (19). Follicular cells, present in the monolayer epithelium, are responsible for iodine uptake and thyroid hormone synthesis. C-cell are intrafollicular or parafollicular cells that are responsible for the production of the calcium-regulating hormone calcitonin (19). The majority of the thyroid tumors (more than 95%) are derived from the follicular cells, while a minority (3%), called medullary thyroid carcinomas, are C-cell derived carcinomas (19). Thyroid tumors are of two types (19).: benign and malignant. Benign tumors are principally represented by adenomas, while malignant tumors are, in most cases, carcinomas. Thyroid carcinomas are one of the most common malignancies of the endocrine system. Follicular cell-derived carcinomas are commonly divided into well-differentiated thyroid carcinoma (WDTC), poorly differentiated thyroid carcinoma (PDTC), and undifferentiated types depending on various histological and clinical factors (19). WDTCs include papillary thyroid carcinoma (PTC) and follicular thyroid carcinoma (FTC) types (reviewed in 19). Some years ago, several studies were undertaken to analyzed the expression of miRNAs in thyroid carcinoma to evaluated a possible role of their deregulation in the process of carcinogenesis (19). These studies showed an aberrant miRNA expression profile that distinguishes uneqivocally among PTC, ATC (anaplastic thyroid cancer), and normal thyroid issue. These studies also demonstrated that miR-221 and miR-222 cluster play a significant role in thyroid carcinoma cell proliferation and that miR-221 and miR-222 are able to impair tumor necrosis factor-related apoptosis inducing ligand (TRAIL)-dependent apoptosis by inhibiting the expression of key functional protein (19). Moreover, the studies indicate that miR-222 plays an important role in cancer cell invasion (19).
OVARIAN CANCER
As mentioned, miRNAs are emerging as important regulators of cancer-related process. A recent study showed that miR-9 is down-regulated in human ovarian cancer relative to normal ovary, and over-expression of miR-9 suppresses cell growth in vitro (121). Furthermore, the 3'UTR of NF-kB1 is found to be regulated directly by miR-9, demonstrating that NF-kB1 is a functionally important target of miR-9 in ovarian cancer cells. When miR-9 is over-expressed in ovarian cancer cells, the mRNA and protein levels of NF-κB1 are both suppressed, whereas inhibition of miR-9 results in an increase the NF-κB1 expression level (121). Ovarian cancer tissues display significantly low expression of miR-9 and a high level of NF-κB1 compared with normal tissues, indicating that regulation of NF-κB1 by miR-9 is an important mechanisms for miR-9 to inhibit ovarian cancer (121).
Most ovarian cancer patients are diagnosed at an advanced stage (67%) and prospects for significant improvement in survival reside in early diagnosis (122). Tumors actively release exosomes into the peripheral circulation and the association of miRNA with circulating tumor-derived exosomes were demonstrated (122). These results suggest that miRNA profiling of circulating tumor exsomes could potentially be used as surrogate diagnostic markers for biopsy profiling, extending its utility to screening asymptomatic populations (122).
PROSTATE CANCER
A recent study showed that miR-15a and miR-16-1 cluster targets encoding cyclin D1(CCND1) and WNT3A, which promotes several tumorigenic features such as survival, proliferation and invasion (123). In cancer cells of advanced prostate tumors, the miR-15a and miR-16 level is significantly decreased, whereas the expression of BCL2, CCND1 and WNT3A is inversely up-regulated (123). Delivery of antagomirs specific for miR-15a and miR-16 to normal mouse prostate results in marked hyperplasia, and knockdown of miR-15a and miR-16 promote survival, proliferation and invasiveness of untransformed prostate cells, which become tumorigenic in immunodeficient NOD-SCID mice (123). Convertsely, reconstitution of miR-15a and miR-16 expression results in growth arrest, apoptosis and marked regression of prostate tumor xenografts (123). The authors proposed that miR-15a and miR-16 act as tumor suppressor genes in prostate cancer through the control of cell survival, proliferation and invasion. These findings have therapeutic implications and may be explored for future treatment of prostate cancer (123). Several miRNAs have been implicated as tumor suppressors based on their physical deletion or reduced expression of human cancer. The miR-15a-16-1 cluster has recently emerged as an excellent candidates for the long sought-after tumor suppressor gene on 13q14 (24,27,68,89,109). The tumor suppressor activity of miR-15a-16-1 is not limited to B cells (2,124). More than 50% of human prostate cancers carry a deletion of 13q14. Accordingly, a recent study has shown that inhibition of miR-15a and miR-16 activity leads to hyperplasia of the prostate in mice and promotes survival, proliferation, and invasion of primary prostate cells in vitro (123). In addition, the therapeutic potential of reconstituting expression of this cluster was illustrated by the significant regression of prostate tumor xenografts upon intra-tumoral delivery of miR-15a and miR-16-1 (90,109). Recently, Zaman et al. investigated the expression and functional significance of miR-145 in prostate cancer. They found that one of the gene significantly up-regulated by miR-145 over-expression is the proapoptotic gene TNFSF 10 (125). Therefore, modulation of miR-145 may be an important therapeutic approach for the management of prostate cancer.
Cancer stem cells (CSCs), or tumor-initiating cells, are involved in tumor progression and metastasis, and miRNAs regulate both normal and CSCs (27). CSCs in many tumors have been identified using the adhesion molecule CD44 (27). A current studies showed that miR-34a, a p53 target, was under-expressed in CD44+ prostate cancer cells purified from xenograft and primary tumors and that enforced expression of miR-34a in bulk or purified CD44+ prostate cancer cells inhibited clonogenic expansion, tumor regeneration, and metastasis (27). In contrast, expression of miR-34a antagomirs in CD44-prostate cancer cells promoted tumor development and metastasis. Interestingly, systemically delivered miR-34a inhibited prostate cancer metastasis and extended survival of tumor-bearing mice, indicating miR-34a is a key negative regulator of CD44+ prostate cancer cells and established a strong rationale for developing miR-34a as a novel therapeutic agent against prostate CSCs (27).
Importantly and interestingly, recent evidence indicates that dietary factors play an important role in the process of prostate carcinogenesis through modulation of miRNA expression (75). Epidemiological data suggest that the rates of clinically significant prostate cancer are 15-fold higher in men from the United States than in men from Asian countries, attributable to the fact that Asian diet comprise a significant portion of soybean-enriched foods (75). Genistein, an isoflavone isolated from soybean, has been found to be a potent antitumor agent which has the capacity to antagonizes estrogen- and androgen-mediated signaling pathways, possesses antioxidant properties, and is a potent inhibitor of angiogenesis and metastasis (75).
HEPATOCELLULAR CARCINOMA (HCC)
Infection of human liver with HBV and HCV induces the development of chronic hepatitis (CH), cirrhosis, and in some instances HCC (126). Ura et al. measured the expression of 188 miRNAs in liver tissues obtained from 12 patients with HBV-related HCC and 14 patients with HCV-related HCC (126). They found that out of the 31 miRNAs associated with disease state, 17 were down-regulated in HCC, which up-regulated cancer-associated pathway such as cell cycle, adhesion, proteolysis, transcription, and translation; 6 miRNAs were up-regulated in HCC, which down-regulated anti-tumor immune response, suggesting miRNAs are important mediators of HBV and HCV infection as well as liver disease progression, and therefore could be potential therapeutic target molecules (126).
LEUKEMIA
miRNA expression profiles can be used to distinguish normal B cells from malignant B cells in patients with CLL. A study showed that a unique miRNA signature is associated with prognostic factors and disease progression in CLL (124). Acute myeloid leukemia (AML) carrying NPM1 mutations and cytoplasmic nucleophosmin (NPMc+AML) accounts for about one-third of adult AML and shows distinct feature, including a unique gene expression profile (112). Garzon et al. investigated the role of miRNA in the biology of NPMc+AML (112). They identified a strong miRNA signature that distinguishes NPMc+mutated from the cytoplasmic-negative (NPM1 unmutated) cases and induces the up-regulation of miR-10a, miR-10b, several let-7 and miR-29 family members and that many of the down-regulated miRNAs including miR-204 and mmiR-128a are predicted to target several Homebox (HOX) gene (112). They also confirmed that miR-204 targets HOXA10 and MEIS1, suggesting that the HOX up-regulation observed in NPMc+AML may be due in part by loss of HOX regulators-miRNAs (112). Further experiments demonstrated that the up-regulation of miR-155 was independent from FLT3 signaling. These results identify a unique miRNA signature associated with NPMc+AML and provide evidence that support a role for miRNAs in the regulation of HOX genes in this leukemia subtype (112). It is known that aberrant miRNA expression can play a vital role in the pathology of leukemia, thus miRNAs have rapidly emerged as potential targets for therapeutics (127). Recent studies showed the important roles of miRNAs in the pathogenesis of leukemia, including AML, acute lymphoblastic leukemia (ALL), chronic myeloid leukemia (CML) and CLL (127).
HODGKIN LYMPHOMA
Currently, Navrro et al. analyzed miRNA expression in classic Hodgkin lymphoma and found that miR-96, miR-128, and miR-128b were selectively down-regulated in classic Hodgkin lymphoma with Epstein-Bar virus, suggesting that miRNAs play an important role in the biology of classic Hodgkin lymphoma and may be useful in developing therapies targeting miRNAs (28).
The discovery of hundreds of distinct regulatory miRNAs has already led to profound changes in our understanding of the genetic control mechanisms that operate in health and disease (65,66). By now, dozens of miRNAs have been reported in various tumor phenotypes as described, and yet these only scratch the surface of the real complexity, as the functions of hundreds of miRNAs known to be present in our cells and altered in expression in different forms of cancer remain total mysteries (66). Here again, we are unclear as to whether future progress will cause fundamental shifts in our understanding of the pathogenetic mechanisms of cancer or only add detail to the elaborate regulatory circuits that have already been mapped out (66).
CARDIOVASCULAR DISEASES
Cardiovascular disease is the predominant cause of human morbidity and mortality in developed countries. Thus, extraordinary effort has been devoted to determining the molecular and pathophysiological characteristics of the diseased heart and vasculature with the goal of developing novel diagnostic and therapeutic strategies to combat cardiovascular disease (28). Recently, miRNAs are implicated in the pathogenesis of various cardiovascular diseases and have become an intriguing target for therapeutic intervention (5,91,128).
Congestive heart failure is a growing problem that reduce life span and lowers the quality of life of those it afflicts. Congestive heart failure is characterized by neurohumoral activation and a decline in cardiac output (129,130). Several miRNAs are regulated uniformly across all causes of heart failure and miRNA regulate embryogenesis and cell fate determination (5,91,128). Without Dicer, mice die in utero ~7.5 days after fertilization (129,131). It has been reported that miRNAs regulate cardiovascular structure and function (5). Interestingly, miRNAs promote the commitment of embryonic stem cells to the cardiomyocyte lineage and cardiomyocyte differentiation and proliferation require temporal and transcription regulation of specific genetic programs (129,131). In addition, miRNA also regulate genes important for ventricular hypertrophy (129). Importantly, the cardiac-specific miRNAs (miR-1, miR-133, and miR-208) participate in the development of ventricular hypertrophy and heart failure (91,129). Over-expression of miR-133 blocked cardiac hypertrophy in Akt transgenic mice (129). The accumulating experimental results clearly show that miRNAs play a significant role in cardiovascular development and disease, and that miRNAs are important for regulating cardiomyocytes self-renewal and differentiation, as well as for normal cardiac structural integrity (128,129,131). Identification of those miRNA and target genes that contribute adult cardiac pathology is likely to suggest new target for therapy.
Atherosclerotic coronary artery disease (CAD) continues to be an important socioeconomic problem and the leading cause of death in the developing countries (29). Major independent risk factors for CAD are known to include familiarity, high blood cholesterol levels, hypertension, smoking, diabetes, and obesity (29). Innovative early and reliable biomarker and therapeutic targets for CAD would thus be acclaimed (29). Recently, miRNAs were found to be present in the circulating bloodstream and it was discovered that plasma and serum miRNAs posses robust stability even after cycles of freezing/thawing (132-135). Moreover, acute myocardial infarction (AMI) is the world's leading cause of morbidity and mortality (6). More recent reports showed that after AMI in humans and mice, muscle-enriched miRNAs, such as miR-1, miR-133a, miR-133b, and miR-499-5p, are increased in plasma and that miR-499 and miR-133a are highly expressed in heart, whereas miR-1 and miR-133a are highly expressed both in heart and in skeletal muscel (6,29). Indicating that circulating miRNAs may become helpful and reliable tools for the diagnosis ad prognosis of patients with CAD or other cardiovascular disease. As mentioned, miR-21 is aberrantly expressed in multiple types of cancer including advanced human breast cancer, cervical and ovarian cancer, colon carcinoma, hepatocellular cancer, glioma, esophageal cancer, B-cell lymphoma, prostate cancer, lung cancer and other cancer types but also in cardiovascular disease (91). The consistent miR-21 overexpression under pathophysiological conditions points to miR-21 as a valuable tool for new therapeutic strategies (91).
Various miRNAs are expressed in a tissue-specific manner, and, thus may regulate tissue-specific functions. Recently, a number of miRNAs have been found to be striated-muscle specific (such as miR-1 and miR-133), whereas only miR-208 has been found to be purely cardiac specific (136). Several recent studies performed microarray analysis to determine whether miRNAs are dysregulated in hypertrophic and failing hearts (5,91,136,137). These studies have revealed signature patterns of miRNAs that are up- and down-regulated during pathological cardiac remodeling in rodents and humans (5,91,136,137). Although miR-1, miR-29, miR-30, miR-133, and miR-150 have often been found to be down-regulated, however, miR-21, miR-23a, miR-125, miR-195, miR-199, and miR-214 are up-regulated with hypertrophy (5,91,136,137). Gain-and loss-of-function studies have implicated these miRNAs in the different aspects of remodeling process during the progression of heart disease (5,137).
The continuous discovery of new miRNAs suggest that the present collection of miRNAs implicated in cardiovascular disease is likely to be incomplete. In diseased hearts, regional changes in electrophysiology can result in nonuniform impulse propagation, which can lead to arrhythmias (5,136). Although arrhythmias usually develop as a result of cardiac disease or inherited gene mutations in ion channel, several miRNAs, including miR-1 and miR-133, have recently been implicated in electophysiological abnormalities (5,136). Ikeda et al. compared miRNA expression in 3 different types of human heart disease (ischemic cardiomyopathy, dilated cardiomyopathy, and aortic stenosis) with normal heart (137). Among the 87 miRNAs detected in the heart, roughly half were differentially expressed in at least 1 disease group, whereas 7 miRNAs were regulated in the same direction in all 3 disease states (137). Especially intriguing is the discovery of a network of muscle-specific miRNAs embedded within myosin heavy chain genes, which control myosin expression and the response of the heart to stress and thyroid hormone signaling (138).
Thioredoxin 1 (Trx1) is a ubiquitously expressed antioxidant that has 2 cystine residues in its catalytic center and one of the most prominent actions of Trx1 in the heart is suppression of cardiac hypertrophy (139). Currently, a study investigated the role of miRNAs in mediating the anti-hypertrophic effect of Trx1 on angiotensin II (AngII)-induced cardiac hypertrophy (139). The authors found that Trx1 up-regulates expression of the let-7 family, including miR-98, which in turn inhibits cardiac hypertrophy, in part through down-regulation of cyclin D2 (139).
AMI involves necrotic and apoptotic loss of cardiomyocytes and one strategy to salvage ischemic cardiomyocytes is to modulate gene expression to promote cell survival without disturbing normal cardiac function (30). Currently, Qian et al. reported that miR-24 expression is down-regulated in the ischemic border zone of the murine left ventricle after AMI (30). The authors also reported that miR-24 suppresses cardiomyocyte apoptosis, in part by direct repression of the BH3-only domain-containing protein Bim, which positively regulates apoptosis and that in vivo expression of miR-24 in a mouse AMI model inhibited cardiomyocyte apoptosis, attenuated infarct size, and reduced cardiac dysfunction (30). Interestingly, this antiapoptotic effect on cardiomyocytes in vivo was partially mediated by Bim (30). These results suggest that manipulating miRNA levels during stress-induced apoptosis may be a novel therapeutic strategy for cardiac disease.
Courboulin et al. also currently reported that miR-204 expression in pulmonary artery smooth muscle cells is down-regulated in both human and rodent pulmonary arterial hypertension (PAH) and that miR-204 down-regulation correlates with PAH severity (140). Importantly, they also reported that delivery of synthetic miR-204 to the lungs of animals with PAH significantly reduced disease severity, suggesting that reestablishing miR-204 expression should be explored as a potential new therapy for this disease. miR-133, which is enriched in cardiac and skeletal muscle, is involved in cell specification, differentiation, and development (141). It is also down-regulated during cardiac hypertrophy, which indicated that it may play a role in the underlying pathogenesis (141).
CONCLUSION REMARKS
Accumulating evidence has demonstrated that microRNAs (miRNAs) play a major role in a wide range of developmental processes including cell proliferation, cell differentiation, cell cycle, metabolism, apoptosis, developmental timing, neuronal cell fate, neuronal gene expression, brain morphogenesis, muscle differentiation and stem cell division. In addition, miRNAs could contribute to pathogenesis of many diseases. Aberrant miRNA function influences human disease. It is interesting to note that some miRNAs may have dual functions as both tumor suppressors and oncogenes. Moreover, a large number of studies have reported link between alteration of miRNA homeostasis and pathological conditions such as psychiatric and neurological diseases as well as cancer, cardiovascular diseases and autoimmune diseases. Furthermore, miRNA deficiencies or excess have been reported to be correlated with a number of clinically important disease ranging from infection and myocardial infarction to cancer. Importantly, relatively small changes in miRNA expression can lead to modest changes in the levels of multiple genes within biological network, and collectively these can add up to large changes in biology. Therefore, it is not surprisingly that miRNAs have been implicated in various human diseases. In this review, I summarized the newly discovered roles of miRNAs in various human cancers and cardiovascular diseases.
In the future, distinct miRNA signature involved in cancers should define the role of miRNA in biochemical and biological processes. Furthermore, identification of miRNAs of significance in disease could provide rationale for the design and implementation of disease classification, early detection, disease prognosis and therapeutic decision making, and other new immunotherapeutic and pharmacological strategies for treatment of diseases. Moreover, miRNAs may represent attractive novel diagnostic biomarkers mainly due to their higher stability when compared to RNAs and could potentially provide possibilities for therapeutic intervention. Unravelling the regulatory circuits of miRNAs in biology is still in its infancy, but will likely yield new insights into our understanding of immunophysiology and immunopathology. It is hoped that research into the normal biochemical process will dovetail with a better understanding of the pathological process that these molecules and pathway play.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download