Abstract
Purpose
Multidrug resistance (MDR) remains a major obstacle in the treatment of triple-negative breast cancer (TNBC) with conventional chemotherapeutic agents. A previous study demonstrated that hsa-miRNA-143-3p plays a vital role in drug resistance of TNBC. Downregulation of hsa-miRNA-143-3p upregulated the expression of its target protein cytokine-induced apoptosis inhibitor 1 (CIAPIN1) in order to activate MDR, while upregulation of hsa-miRNA-143-3p effectively enhances the sensitivity of drug-resistant TNBC cells to chemotherapeutics. The present study aimed to further verify these findings in vivo.
Methods
We established a hypodermic tumor nude mice model using paclitaxel-resistant TNBC cells. We expressed ectopic hsa-miRNA-143-3p under the control of a breast cancer-specific human mammaglobin promoter that guided the efficient expression of exogenous hsa-miRNA-143-3p only in breast cancer cells. Thereafter, we overexpressed hsa-miRNA-143-3p in xenografts using a recombinant virus system and quantified the expression of hsa-miRNA-143-3p, CIAPIN1 protein, and proteins encoded by related functional genes by western blot.
Results
We successfully completed the prospective exploration of the intravenous virus injection pattern from extensive expression to targeted expression. The overexpression of hsa-miRNA-143-3p significantly alleviated chemoresistance of TNBC by inhibiting viability. In addition, we observed that the expression of CIAPIN1 as a hsa-miRNA-143-3p target protein was remarkably decreased.
Triple-negative breast cancer (TNBC) is a type of invasive primary breast carcinoma that lacks expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) [1]. TNBC is not sensitive to endocrine therapy or HER2-targeted therapy owing to the lack of antigens, so chemotherapy is the standard treatment. A therapeutic schedule based on paclitaxel remains standard [2], whereas patients with TNBC exhibiting multidrug resistance (MDR) are less sensitive to chemotherapy, which severely weakens the clinical effect [3]. Previous studies show that an important resistance mechanism is the overexpression of a P-glycoprotein encoded by the MDR1 gene as an efflux pump in paclitaxel-resistant tumor cells [4]. Other mechanisms include mutation of the paclitaxel binding protein tubulin, deregulation of cell cycle, and high expression of anti-apoptotic protein [5]. Therefore, novel strategies to reverse MDR in TNBC need to be urgently explored.
MicroRNAs (miRNAs) are endogenous noncoding small RNAs that consist of 21 to 22 nucleotides that are abundant in organisms. miRNAs mainly bind to 3′ untranslated regions (3′-UTRs) of their target mRNAs, specifically via complete or incomplete matching to miRNA recognition element sequences, thereby inducing the degradation of the target mRNA, inhibiting translation, or regulating gene transcription [6]. An increasing number of studies suggest that the expression pattern of miRNAs plays a vital role in drug resistance and is involved in drug metabolism and distribution through regulating drug metabolism enzymes, drug transport molecules, transcription factors or nucleotide receptors, or other mechanisms [7]. In recent years, hsa-miRNA-143-3p has attracted tremendous attention as a new player in drug resistance in tumors. For example, this miRNA can enhance doxorubicin toxicity in osteosarcoma cells by inhibiting the expression of autophagy related 2 homolog B (ATG2B), B-cell lymphoma-2 (Bcl-2), and autophagy marker light chain 3 (Lc3-I) [8] and enhances the apoptosis effect induced by platinum in gastric cancer through regulating insulin-like growth factor 1-receptor (IGF-1R) and Bcl-2 [9], as well as participating in MDR of colorectal cancer [10] and bladder cancer [11]. However, the role of this miRNA has not been explored in TNBC.
In 2004, cytokine-induced apoptosis inhibitor 1 (CIAPIN1) was identified as a newly discovered antiapoptotic protein [12]. Subsequent studies indicated that this protein has important functions in MDR development in multiple tumor types, such as colon cancer [13], gastric cancer [14], leukemia [15], and breast cancer [16]. Surprisingly, in a preliminary in vitro study, we found that hsa-miRNA-143-3p has the CIAPIN1 gene mRNA 3′-UTR seed binding site in TNBC cells [17], which suggests that CIAPIN1 may be a target of hsamiRNA-143-3p.
In the present study, we introduced exogenous hsa-miRNA-143-3p using a recombinant lentivirus vector containing the breast cancer human mammaglobin (hMAM) gene promoter harboring high transcription activity and specificity, aiming to specifically increase the expression level of hsa-miRNA-143-3p in TNBC xenografts. Simultaneously, we showed that high expression of hsa-miRNA-143-3p inhibited the growth of drug-resistant tumors and decreased the expression of the CIAPIN1 protein. Hence, we clarified that hsa-miRNA-143-3p could enhance paclitaxel sensitivity in drug-resistant TNBC by affecting the expression of CIAPIN1.
BT-20, MDA-MB-231, MCF7, SKBr-3, Lovo, HepG2A549, 293T, and MRC-5 cells were obtained from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). MDA-MB-231/Tax cell lines were established in a previous experiment [17] and passaged with trypsin digestion. MDA-MB-231/Tax cell lines in logarithmic growth phase at passage 3 were prepared into cell suspensions at 1×108/mL and preserved at 4℃. All cells were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, USA) containing 10% fetal bovine serum (Invitrogen) and maintained in a humidified 5% CO2 incubator at 37℃.
Approximately 5×106 MRC-5 cells (CAS, Beijing, China) in logarithmic phase were collected. The Qiagen genomic DNA extraction kit (Qiagen, Shanghai, China) was used to extract genomic DNA. Genomic DNA (500 ng) was used as a template to amplify the hMAM gene promoter sequence by polymerase chain reaction (PCR). Primers for hMAM promoter were: 5′-GCTCTAGA AGTTTTAATCCTGCTGCAAACCAG-3′ (forward) and 5′-GCTCTAGA ATTAAGAAAAGCACACTGGCGAATGAAC-3′ (reverse). The cycling conditions were as follows: 35 cycles of denaturation at 94℃ for 30 seconds, annealing at 55℃ for 30 seconds, and extension at 72℃ for 40 seconds. The PCR product was inserted into the recombinant expression vector pcDH-CMV-miRNA-143-3p to construct a specific promoter expression vector called the pcDH-hMAM-vector by replacing the original CMV promoter. The recombinant vector was identified by sequence analysis. 293TN cells (System Biosciences, Johnstown, USA) were transfected with virus packaging plasmid mixtures to produce recombinant viruses named Lv-miRNA-143-3p and Lv-hMAM-miRNA-143-3p, respectively.
To investigate the expression efficiency of the hMAM gene promotor, MDA-MB-231 cells were thawed and cultured. Briefly, cells in logarithmic growth phase were seeded in 6-well plates and transfected with recombinant virus named definited no-antisense oligomers control (NC), Lv-miRNA-143-3p, and Lv-hMAM-miRNA-143-3p after overnight culture. A blank control group was also examined. Seventy-two hours after infection, cells were collected. TRIzol reagent (Invitrogen) was used to extract total RNA according to the manufacturer's protocols. The levels of mature hsa-miRNA-143-3p mRNA were measured using SYBR Green incorporation on a Roche LightCycler480 Real Time PCR system (Roche Diagnostics, Rotkreuz, Switzerland), with U6 expression as an internal control.
To investigate the expression specificity, cells groups for BT-20, MDA-MB-231, MCF7, SKBr-3, Lovo, HepG2, and A549 (Shanghai Institute of Cell Biology) in logarithmic growth phase were cultured and seeded in 6-well plates. Simultaneously, cells in each group were infected with Lv-NC, Lv-miRNA-143-3p, or Lv-hMAM-miRNA-143-3p after overnight culture. A blank control group was also examined. After infection for 72 hours, cells were collected to extract total RNA. The levels of mature hsa-miRNA-143-3p mRNA were equal among groups.
Female athymic BALB/c nude mice (n=24; weight, 18–20 g) were obtained from Shanghai Sippr-BK Laboratory Animal Co., Ltd. (Shanghai, China). All animal experiments were approved by the Ethics Committee at Harbin Medical University. Briefly, a total of 0.5 mL MDA-MB-231/Tax cells were subcutaneously injected into the armpits of the left anterior limb of each mouse and then monitored every other day for tumor growth. When the tumor grew to 2.5×2.5 mm2 on the 12th day, the mice were randomly divided into four experimental groups called control, paclitaxel, paclitaxel+Lv-NC, and paclitaxel+Lv-hMAM-miRNA-143-3p groups.
Fifteen days after inoculating MDA-MB-231/Tax cells, mice in the experimental groups were respectively injected with Lv-hMAM-miRNA-143-3p or Lv-NC through the tail vein at 50 µL (1×107 ifu/µL) for 6 times in 3 days. Paclitaxel (6 µg/µL; Corden Pharma Latina S.p.A., Sermoneta, Italy) was administered at 20 mg/kg at 24 hours after the final virus injection twice a week for 4 weeks based on the half maximal inhibitory concentration of MDA-MB-231/Tax cells determined by our previous trial. Meanwhile, mice in the control groups were given equal volumes of saline. Tumor size was measured at 1, 2, 3, and 4 weeks after the first dose to acquire tumor volume proliferation curves. The tumor volume was calculated as (short diameter2×long diameter)/2. After treatment for 5 weeks, the tumor tissue was separated, weighed, and finally preserved in liquid nitrogen. The tumor growth inhibition value was used to test the inhibitory effect of drugs on tumor growth. The value of inhibition ratio=(1−mean weight value of experimental group/mean weight value of control group)×100%.
Frozen tissues were dissociated by TRIzol (Invitrogen) and then total RNA was isolated, extracted, and its concentration and quality were determined. Reverse transcription to synthesize cDNA was accomplished by using reverse transcriptase M-MLV (Takara, Dalian, China). The real-time PCR primers of hsa-miRNA-143-3p were as follows: forward, 5′-TGAGATGAAGCACTGTAGCTC-3′ and reverse, 5′-GTCGTATCCAGTGCGTGTCGTG-3′. U6 (H. sapiens snRNA) was used as an internal control. The cycling conditions were as follows: 45 cycles of denaturation at 95℃ for 10 seconds, annealing at 55℃ for 10 seconds, and extension at 72℃ for 10 seconds. In a Relative Quantification test, the data were analyzed using the cycle threshold (Ct) value obtained from Thermal Cycler DICE Real Time System analysis software (TakaRa, Kyoto, Japan) using the intersection method compared with β-actin mRNA levels. The relative quantitative results were parsed by the 2ΔΔCt method and the expression quantity of target gene relative to the endogenous reference was 2ΔCt=2Ctm−Ctn.
Total proteins were extracted from tissues using T-PER Tissue Protein Extraction Reagent (Pierce, Rockford, USA). Protein concentrations were determined using a BCA Protein Assay Kit (Pierce). Aliquots of tissues extract containing 100 µg (10 µg/µL) of total protein were resolved using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Millex-Hv 0.45 mm polyvinylidene fluoride filter membranes (Millipore, Burlington, USA). Membranes were then blocked in 5% dried skim milk in Tris buffered saline with Tween-20 (TBST) (8 g NaCl, 2.42 g Tris-HCl, 0.5% Tween-20) for 2 hours at room temperature (RT; 20℃) and then incubated at 4℃ overnight with anti-CIAPIN1 (Abcam, Cambridge, UK), anti-P53 (Abcam), anti-P-gp (Abcam), or anti-glyceraldehydephosphate dehydrogenase (GAPDH) monoclonal antibodies (Abcam). After three washes in TBST buffer (10 minutes, RT), the membranes were incubated for 1 hour at RT with an anti-mouse (Abcam) or anti-rabbit (Abcam) secondary antibody as appropriate. After three washes in TBST buffer (10 minutes, RT), the enhanced chemiluminescence luminol reagent (Pierce) was used to develop the membrane according to the manufacturer's recommendation. GAPDH protein was used as a loading control. All procedures were performed three times.
In order to verify the efficiency and specific expression of the hMAM promoter in TNBC cells, we detected the expression efficiency of hsa-miRNA-143-3p under the control of different promoters in TNBC cells and the expression level of hsa-miRNA-143-3p guided by the hMAM promoter in different tumor cells, respectively. Data indicated that after transfer with Lv-NC, Lv-miRNA-143-3p and Lv-hMAM-miRNA-143-3p for 72 hours in MDA-MB-231 cells, The content of hsa-miRNA-143-3p in cells infected with Lv-miRNA-143-3p and Lv-hMAM-miRNA-143-3p was significantly higher than that in the control group and virus infection control group (p<0.01). Although the expression of hsa-miRNA-143-3p in the Lv-hMAM-miRNA-143-3p infection group was slightly higher than that in the Lv-miRNA-143-3p infection group, there was no significant difference (p>0.05), suggesting that the hMAM promoter can effectively guide the transcription of hsa-miRNA-143-3p in MDA-MB-231 breast cancer cells (Figure 1). In addition, expression specificity data showed that after infecting MDA-MB-231, MCF7, BT-20, SKBr-3, Lovo, HepG2, and A549 cells with Lv-hMAM-miRNA-143-3p for 72 hours respectively, the content of hsa-miRNA-143-3p in BT-20, MDA-MB-231, MCF7, and SKBr-3 cells was significantly higher than that of the cells in the control group and the Lv-NC infection group (p<0.01). In Lovo, HepG2, and A549 cells infected with Lv-hMAM-miRNA-143-3p, the content of intracellular hsa-miRNA-143-3p was slightly higher than that in the cells in control and Lv-NC infection groups, but there was no significant difference (p>0.01). Meanwhile, the expression was statistically significantly higher in the Lv-miRNA-143-3p infection group than expression in the Lv-hMAM-miRNA-143-3p infection group, Lv-NC infection group, and control group (p<0.01). Cells in each group were infected with Lv-hMAM-miRNA-143-3p for 72 hours. In BT-20, MDA-MB-231, MCF7, and SKBr-3 breast cancer cells lines, the content of hsa-miRNA-143-3p was significantly higher than that in Lovo, HepG2, and A549 cells (p<0.01), indicating that the hMAM promoter had higher transcriptional activity in TNBC cells than in other tumor types (Figure 1).
In order to elucidate the effect of hsa-miRNA-143-3p on the regulation of MDR in TNBC, we compared the subcutaneous tumor proliferation curves of each group to analyze the drug sensitivity of each treatment group to paclitaxel. The results showed that the tumor volume growth rate and tumor size in the Lv-hMAM-miRNA-143-3p+paclitaxel group were significantly reduced compared with those in the Lv-NC+paclitaxel group, paclitaxel group, and physiological saline control group (Figure 2A).
Upregulating the expression of hsa-miRNA-143-3p statistically significantly inhibited tumor growth (p<0.01) (Figure 2B). Our findings indicated that hsa-miRNA-143-3p expression could improve sensitivity to paclitaxel in TNBC.
By quantifying the expression of hsa-miRNA-143-3p and CIAPIN1 protein, we conducted inductive analysis of the hsamiRNA-143-3p/CIAPIN1 drug resistance pathway in vivo. Via real-time PCR detection, we found that the RNA expression level of hsa-miRNA-143-3p in the Lv-hMAM-miRNA-143-3p+paclitaxel group was remarkably increased compared with its expression in the Lv-NC+paclitaxel group, paclitaxel group, and physiological saline control group (p<0.01) (Figure 3A). Accordingly, western blot detection results indicated a statistically significant decrease of the expression level of CIAPIN1 protein (p<0.01) (Figure 3B). These data confirmed that hsa-miRNA-143-3p affected the expression of its target CIAPIN1 protein.
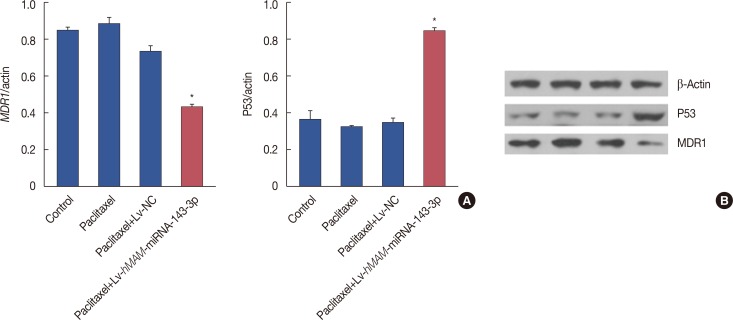
To further elucidate the influence of hsa-miRNA-143-3p on MDR in TNBC, we examined changes in the protein expression level of the drug resistance-related proteins P53 and MDR1 the after gene intervention in the tumors by western blot. As expected, the expression of P53 protein increased while expression of MDR1 protein decreased in the Lv-hMAM-miRNA-143-3p+paclitaxel group, compared with that in the Lv-NC+paclitaxel group, paclitaxel group, and physiological saline control group, and both these effects were statistically significant (p<0.01) (Figure 4).
MDR remains a major obstacle to achieving curative breast cancer chemotherapy treatment, and causes >90% of metastatic breast cancer patients to experience treatment failure [3]. Structural variations and dysregulation of miRNAs have been shown to affect the formation of mature miRNAs and also MDR [18]. Abnormal expression of miRNAs participates in MDR through regulation of protein expression of chemosensitive or chemoresistant genes [19]. It has been reported that miR-761 promotes the sensitivity of colorectal cancer cells to 5-fluorouracil through targeting FOXM1 [20]. Another study showed that miR 647 overexpression led to decreased migration and invasion of SGC 7901/VCR cells (vincristine-resistant gastric cancer cells) by targeting ankyrin-B (ANK2), focal adhesion kinase (FAK), matrix metalloproteinase (MMP2 and MMP12), cluster of differentiation (CD44), and snail family transcriptional repressor 1 (SNAIL1) [21]. In addition, overexpression of miR-216b sensitizes non-small cell lung cancer cells to cisplatin-induced apoptosis by targeting c-Jun [22]. Restoring miR-27b levels was reported to enhance response to paclitaxel by directly targeting casitas B cell lymphoma-b and growth factor receptor-bound protein 2 in breast cancer [18]. Taken together, the results of these studies strongly suggest miRNAs as promising targets to combat drug resistance.
A preliminary in vitro study highlighted the significance of hsa-miRNA-143-3p, finding that its expression levels were negatively correlated with paclitaxel resistance in TNBC and upregulating its expression could notably strengthen the ability of paclitaxel to kill tumor cells [17]. This previous study first demonstrated that the expression profile of hsa-miRNA-143-3p could provide MDR information and also acted as a reserve force to reverse MDR. Owing to the close relation between the emergence of tumor resistance and the tumor microenvironment, as well as the intratumoral heterogeneity and complexity, we sought further verification using drug-resistant TNBC xenografts as an in vivo model of chemoresistance seen in the clinic. First, we needed to introduce ectogenic hsamiRNA-143-3p with a recombinant lentivirus that leads to efficient transfection and sustained expression of the desired gene to host cells [23]. Regrettably, relying on insertional mutagenesis of lentivirus-expression vectors to induce oncogenesis poses a safety risk and may cause abnormal expression of the transgene in adjacent normal tissues [24], therefore, it is crucial to ensure correct targeting of the recombinant lentivirus. A previous study confirmed that hMAM, a breast cancerspecific endocrine globulin, can control specific expression to breast tissue [25]. We compared the expression efficiency of different promoters in TNBC cells including the known efficient CMV promoter and the hMAM promoter in different tumor cells, respectively, to assess the high transcription activity and specificity of the hMAM promoter in TNBC. Correspondingly, we integrated the hMAM promoter into Lv-miRNA-143-3p to guide hsa-miRNA-143-3p positioning to tumor tissue. This exploration has preliminarily switched the pattern of gene intervention from extensive to targeted. Next, we identified that overexpression of hsa-miRNA-143-3p limited tumor growth and improved sensitivity after undergoing gene interference through analyzing tumor survival curves. Taken together, our results provided forceful preclinical support that hsa-miRNA-143-3p should be pivotal target of reversing MDR in TNBC.
Notably, the current study found that the expression of CIAPIN1 was downregulated after upregulating hsa-miRNA-143-3p in TNBC xenografts. Previous bioinformatics prediction studies and literature searches found that hsa-miRNA-143-3p highly matches the 3′-UTR site of the CIAPIN1 gene. Intriguingly, overexpression of hsa-miRNA-143-3p decreased the expression of CIAPIN1 to inhibit cell viability in vitro. These observations commonly suggested that CIAPIN1 might be advantageous for tumors and chemotherapy-induced resistance targeted by hsa-miRNA-143-3p. Increasing studies have revealed that P53 inhibits tumor cell proliferation in association with apoptosis signaling, and MDR1, an adenosine triphosphate combined transporter (ABC), triggers drug efflux. Both these proteins showed the most relevance to tumor MDR [2627]. The results of the present study relying on the increasing expression of P53 and the decrease of MDR1 supported the mechanism for regulation of hsa-miRNA-143-3p to MDR, laying the foundation to the further examine the hsa-miRNA-143-3p/CIAPIN1 pathway. In summary, our results potently validated that hsa-miRNA-143-3p alleviated MDR in TNBC by downregulating the expression level of CIAPIN1.
Recently, the mechanisms by which miRNA regulate their target genes have undergone scrutiny; regulation mainly occurs via negative control at the posttranscriptional level by binding to the 3′- or 5′-UTR region of their target mRNA [28]. Nevertheless, there remains many aspects of MDR that involve miRNA, such as apoptosis, DNA damage repair pathways, manipulation of cell cycle, drug efflux transporters, epithelial-mesenchymal transformation, and tumor microenvironment [29], reflecting that these regulatory mechanisms interplay with each other rather acting alone. Although the present study lacks a deep analysis of the molecular signal transduction mechanism of the hsa-miRNA-143-3p/CIAPIN1 pathway, we will elaborate on this in future studies.
In summary, this study demonstrated that restoring hsamiRNA-143-3p expression facilitates the reversal of MDR by affecting the expression of its target protein CIAPIN1. Precisely targeted gene interventions of hsa-miRNA-143-3p exhibits broad prospects to treat chemoresistant TNBC in the future.
References
1. Zhou S, Sun X, Yu L, Zhou R, Li A, Li M, et al. Differential expression and clinical significance of epithelial-mesenchymal transition markers among different histological types of triple-negative breast cancer. J Cancer. 2018; 9:604–613. PMID: 29483966.

2. Yardley DA, Arrowsmith ER, Daniel BR, Eakle J, Brufsky A, Drosick DR, et al. TITAN: phase III study of doxorubicin/cyclophosphamide followed by ixabepilone or paclitaxel in early-stage triple-negative breast cancer. Breast Cancer Res Treat. 2017; 164:649–658. PMID: 28508185.

3. Doddapaneni R, Patel K, Chowdhury N, Singh M. Reversal of drug-resistance by noscapine chemo-sensitization in docetaxel resistant triple negative breast cancer. Sci Rep. 2017; 7:15824. PMID: 29158480.

4. Vaidyanathan A, Sawers L, Gannon AL, Chakravarty P, Scott AL, Bray SE, et al. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br J Cancer. 2016; 115:431–441. PMID: 27415012.

5. Yang CH, Wang C, Ojima I, Horwitz SB. Taxol analogues exhibit differential effects on photoaffinity labeling of beta-tubulin and the multidrug resistance associated P-glycoprotein. J Nat Prod. 2018; 81:600–606. PMID: 29517223.
6. Ramassone A, Pagotto S, Veronese A, Visone R. Epigenetics and microRNAs in cancer. Int J Mol Sci. 2018; 19:pii: E459.

7. Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017; 141:220–230. PMID: 28240776.

8. Zhou J, Wu S, Chen Y, Zhao J, Zhang K, Wang J, et al. MicroRNA-143 is associated with the survival of ALDH1+CD133+ osteosarcoma cells and the chemoresistance of osteosarcoma. Exp Biol Med (Maywood). 2015; 240:867–875. PMID: 25576341.

9. Zhuang M, Shi Q, Zhang X, Ding Y, Shan L, Shan X, et al. Involvement of miR-143 in cisplatin resistance of gastric cancer cells via targeting IGF1R and BCL2. Tumour Biol. 2015; 36:2737–2745. PMID: 25492481.

10. Simmer F, Venderbosch S, Dijkstra JR, Vink-Börger EM, Faber C, Mekenkamp LJ, et al. MicroRNA-143 is a putative predictive factor for the response to fluoropyrimidine-based chemotherapy in patients with metastatic colorectal cancer. Oncotarget. 2015; 6:22996–23007. PMID: 26392389.

11. Wang H, Li Q, Niu X, Wang G, Zheng S, Fu G, et al. miR-143 inhibits bladder cancer cell proliferation and enhances their sensitivity to gemcitabine by repressing IGF-1R signaling. Oncol Lett. 2017; 13:435–440. PMID: 28123579.

12. Huang Z, Su GF, Hu WJ, Bi XX, Zhang L, Wan G. The study on expression of CIAPIN1 interfering hepatocellular carcinoma cell proliferation and its mechanisms. Eur Rev Med Pharmacol Sci. 2017; 21:3054–3060. PMID: 28742201.
13. Zhang YF, Li XH, Shi YQ, Wu YY, Li N, He Q, et al. CIAPIN1 confers multidrug resistance through up-regulation of MDR-1 and Bcl-L in LoVo/Adr cells and is independent of p53. Oncol Rep. 2011; 25:1091–1098. PMID: 21240465.

14. Zhang XW, Liu L, Zhang XZ, Bo P. Kanglaite inhibits the expression of drug resistance genes through suppressing PVT1 in cisplatin-resistant gastric cancer cells. Exp Ther Med. 2017; 14:1789–1794. PMID: 28810651.

15. Wang J, Li Q, Wang C, Xiong Q, Lin Y, Sun Q, et al. Knock-down of CIAPIN1 sensitizes K562 chronic myeloid leukemia cells to Imatinib by regulation of cell cycle and apoptosis-associated members via NF-kappaB and ERK5 signaling pathway. Biochem Pharmacol. 2016; 99:132–145. PMID: 26679828.
16. Wang XM, Gao SJ, Guo XF, Sun WJ, Yan ZQ, Wang WX, et al. CIAPIN1 gene silencing enhances chemosensitivity in a drug-resistant animal model in vivo. Braz J Med Biol Res. 2014; 47:273–278. PMID: 24676475.

17. Wang JH, Wang XW, Qu D, Sun JW, Guo FX, Lu D. Upregulation of microRNA-143 reverses drug resistance in human breast cancer cells via inhibition of cytokine-induced apoptosis inhibitor 1. Oncol Lett. 2017; 13:4695–4700. PMID: 28588724.

18. Chen D, Si W, Shen J, Du C, Lou W, Bao C, et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018; 9:188. PMID: 29416005.

19. Kopczyńska E. Role of microRNAs in the resistance of prostate cancer to docetaxel and paclitaxel. Contemp Oncol (Pozn). 2015; 19:423–427. PMID: 26843836.

20. Cao S, Lin L, Xia X, Wu H. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-fluorouracil through targeting FOXM1. Oncotarget. 2017; 9:321–331. PMID: 29416616.

21. Cao W, Wei W, Zhan Z, Xie D, Xie Y, Xiao Q. Regulation of drug resistance and metastasis of gastric cancer cells via the microRNA647-ANK2 axis. Int J Mol Med. 2018; 41:1958–1966. PMID: 29328428.

22. Huang G, Pan J, Ye Z, Fang B, Cheng W, Cao Z. Overexpression of miR-216b sensitizes NSCLC cells to cisplatin-induced apoptosis by targeting c-Jun. Oncotarget. 2017; 8:104206–104215. PMID: 29262633.

23. Sharon D, Kamen A. Advancements in the design and scalable production of viral gene transfer vectors. Biotechnol Bioeng. 2018; 115:25–40. PMID: 28941274.

24. Lukashev AN, Zamyatnin AA Jr. Viral vectors for gene therapy: current state and clinical perspectives. Biochemistry (Mosc). 2016; 81:700–708. PMID: 27449616.

25. Li C, Zhang T. Human mammaglobin: a specific marker for breast cancer prognosis. J BUON. 2016; 21:35–41. PMID: 27061528.
26. Xue C, Wang C, Sun Y, Meng Q, Liu Z, Huo X, et al. Targeting P-glycoprotein function, p53 and energy metabolism: combination of metformin and 2-deoxyglucose reverses the multidrug resistance of MCF-7/Dox cells to doxorubicin. Oncotarget. 2017; 8:8622–8632. PMID: 28052008.

27. Gameiro M, Silva R, Rocha-Pereira C, Carmo H, Carvalho F, Bastos ML, et al. Cellular models and in vitro assays for the screening of modulators of P-gp, MRP1 and BCRP. Molecules. 2017; 22:pii: E600.

28. Yang G, Xiong G, Cao Z, Zheng S, You L, Zhang T, et al. miR-497 expression, function and clinical application in cancer. Oncotarget. 2016; 7:55900–55911. PMID: 27344185.

29. Xiong G, Feng M, Yang G, Zheng S, Song X, Cao Z, et al. The underlying mechanisms of non-coding RNAs in the chemoresistance of pancreatic cancer. Cancer Lett. 2017; 397:94–102. PMID: 28254409.

Figure 1
Detection of expression efficiency and specificity of hsa-miRNA-143-3p under control of the hMAM promoter. (A) The recombinant viruses were Lv-NC, Lv-miRNA-143-3p (containing the CMV promoter), and Lv-hMAM-miRNA-143-3p (containing the hMAM promoter). For 72 hours after infection of MDA-MB-231 cells, the relative content of hsa-miRNA-143-3p is shown, with U6 level as a reference. Relative hsa-miRNA-143-3p values were normalized to that of the control group as a reference for data comparison. (B) MDA-MB-231, BT-20, MCF7, SKBr-3, Lovo, HepG2, and A549 cells were infected with Lv-NC, Lv-miRNA-143-3p, or Lv-hMAM-miRNA-143-3p, respectively. After 72 hours, the relative content of hsa-miRNA-143-3p was determined, with U6 level as a reference. Relative hsa-miRNA-143-3p values were normalized to that of the control group as a reference for data comparison. The experimental design was repeated 3 times. Data are represented as mean±standard error of the mean.
Lv-hMAM-miRNA-143-3p=lentiviral-expressed vector human mammaglobin (hMAM) miRNA-143-3p; Lv-NC=lentiviral-expressed vector no-antisense oligomers control group; CMV=cytomegalovirus. *p=0.001 vs. control group.
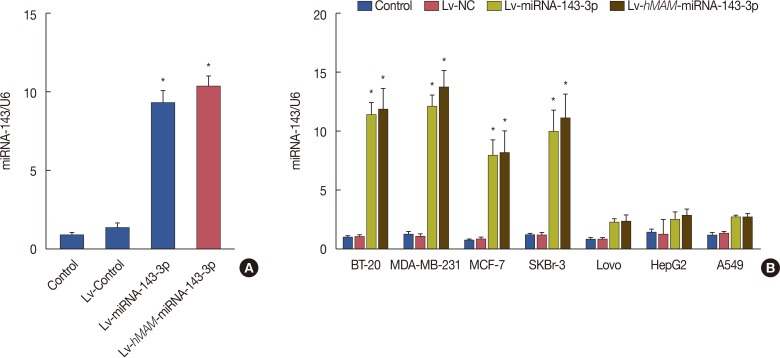
Figure 2
Overexpression of hsa-miRNA-143-3p and the sensitivity of triple-negative breast cancer to paclitaxel. (A) The tumor volume growth rate and tumor size in the Lv-hMAM-miRNA-143-3p+paclitaxel group were significantly reduced compared with those in the Lv-NC+paclitaxel group, paclitaxel group, and physiological saline control group. (B) Upregulating the expression of hsa-miRNA-143-3p prominently increased the tumor growth inhibition ratio in the Lv-hMAM-miRNA-143-3p+paclitaxel group compared to that of the other groups. The experimental design was repeated 3 times. Data are represented as mean±standard error of the mean.
Lv-hMAM-miRNA-143-3p=lentiviral-expressed vector human mammaglobin (hMAM) miRNA-143-3p; Lv-NC=lentiviral-expressed vector no-antisense oligomers control group. *p=0.001.
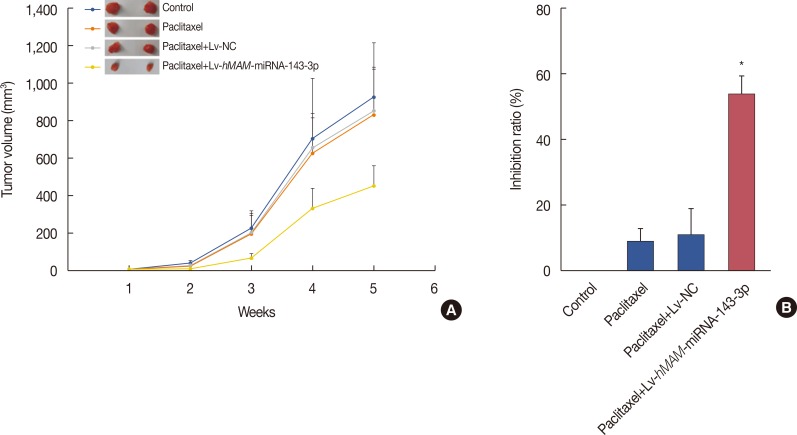
Figure 3
Activation of multidrug resistance by cytokine-induced apoptosis inhibitor 1 (CIAPIN1) targeting to hsa-miRNA-143-3p in triple-negative breast cancer (TNBC). (A) Transcript level of hsa-miRNA-143-3p notably increased upon infection with hsa-miRNA-143-3p compared with the level in other groups as determined by real-time polymerase chain reaction. (B) Western blot showing that CIAPIN1 protein levels decreased in TNBC tissue expressing hsa-miRNA-143-3p. The experimental design was repeated 3 times. Data are represented as mean±standard error of the mean.
β-actin=reference protein; Lv-hMAM-miRNA-143-3p=lentiviral-expressed vector hMAM miRNA-143-3p; Lv-NC=lentiviral-expressed vector no-antisense oligomers control group. *p=0.001.
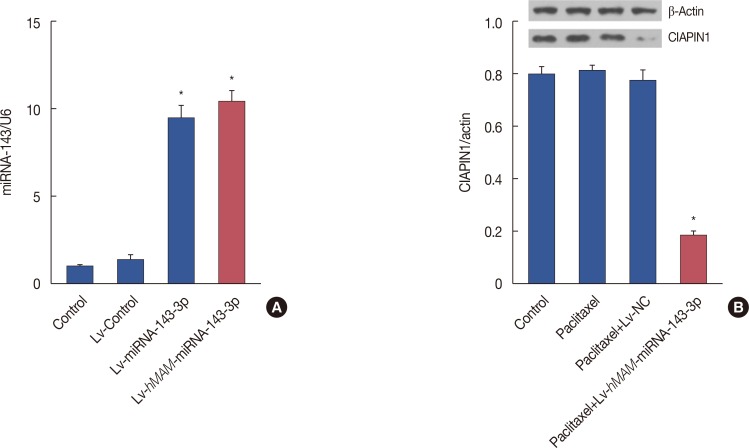
Figure 4
Hsa-miRNA-143-3p expression and other drug resistance-relating proteins. Western blot showing that P53 protein levels increased while multidrug resistance gene 1 (MDR1) levels decreased in the Lv-hMAM-miRNA-143-3p+paclitaxel group compared to levels in other groups. The experimental design was repeated 3 times. Data are represented as mean±standard error of the mean. (A) Western blot showing that P53 protein levels increased while MDR1 levels decreased in the Lv-hMAM-miRNA-143-3p+paclitaxel group compared to levels in other groups. (B) Electrophoretic band of the P53 and MDR1 expression level as described above.
β-actin=reference protein; Lv-hMAM-miRNA-143-3p=lentiviral-expressed vector hMAM miRNA-143-3p; Lv-NC=lentiviral-expressed vector no-antisense oligomers control group. *p=0.001.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download