Abstract
Background
A disputed rpoB mutation is a specific type of rpoB mutation that can cause low-level resistances to rifampin (RIF). Here, we aimed to assess the frequency and types of disputed rpoB mutations in Mycobacterium tuberculosis isolates from South Korea.
Methods
Between August 2009 and December 2015, 130 patients exhibited RIF resistance on the MTBDRplus assay at Asan Medical Center. Among these cases, we identified the strains with disputed rpoB mutation by rpoB sequencing analysis, as well as among the M. tuberculosis strains from the International Tuberculosis Research Center (ITRC).
Results
Among our cases, disputed rpoB mutations led to RIF resistance in at least 6.9% (9/130) of the strains that also exhibited RIF resistance on the MTBDRplus assay. Moreover, at the ITRC, sequencing of the rpoB gene of 170 strains with the rpoB mutation indicated that 23 strains (13.5%) had the disputed mutations. By combining the findings from the 32 strains from our center and the ITRC, we identified the type of disputed rpoB mutation as follows: CTG511CCG (L511P, n=8), GAC516TAC (D516Y, n=8), CTG533CCG (L533P, n=8), CAC526CTC (H526L, n=4), CAC526AAC (H526N, n=3), and ATG515GTG (M515V, n=1).
Isoniazid (INH) and rifampin (RIF) are the most important drugs administered for the treatment of tuberculosis (TB). Resistance to both of these drugs results in multidrug-resistant (MDR) TB, which is a major threat to TB control in South Korea1. RIF resistance is rare among INH-susceptible strains, as the mutation to develop spontaneous INH resistance occurs in approximately one in every 106
Mycobacterium tuberculosis bacilli present; in contrast, the mutation rate is one in every 108
M. tuberculosis bacilli for RIF2. Mutations in the well-defined 81-bp region of the rpoB gene, known as the RIF-resistance-determining region (RRDR), have been detected in approximately 97% of all RIF-resistant M. tuberculosis strains3.
The degree of resistance among the mutations in the RRDR region is not equal. In particular, some RRDR mutations—called disputed rpoB mutations—can cause low-level resistance to RIF. A previous report defined low-level resistance to RIF as a minimal inhibitory concentration (MIC) of 0.063–0.5 µg/mL for RIF using the 7H9 Middlebrook medium4. M. tuberculosis isolates with the disputed rpoB mutation exhibit discrepant susceptibility results of RIF resistance between genotypic and phenotypic tests5. That is, the phenotypic drug susceptibility test (DST) may indicate susceptibility to RIF, despite genotypic resistance to RIF. The commonly observed disputed rpoB mutations in previous studies include CTG511CCG (L511P)4, GAC516TAC (D516Y)6, CAC526CTC (H526L)7, and CTG533CCG (L533P)5. In contrast, undisputed rpoB mutations, including GAC516GTC (D516V), CAC526TAC (H526Y), CAC526GAC (H526D), and TCG531TTG (S531L), are known to cause high-level RIF resistance89
Disputed rpoB mutations have previously been considered to be very rare10. However, a recent report suggested that they frequently contribute to RIF resistance7. To our knowledge, no studies on disputed rpoB mutations have been conducted in South Korea thus far. Therefore, in the present study, we aimed to investigate the frequency and types of disputed rpoB mutations, as well as the treatment outcomes for patients with M. tuberculosis isolates with disputed rpoB mutations.
The study was conducted at the Asan Medical Center, which is a 2,700-bed referral hospital in Seoul, South Korea (an intermediate TB burden country). A total of 1,551 Genotype MTBDRplus assays (Hain Lifescience, Nehren, Germany), one of the World Health Organization (WHO) endorsed line probe assays11, were performed between August 2009 and December 2015. Of the 1,389 MTBDRplus assays from which corresponding phenotypic DST results were available, 130 (9.4%) exhibited RIF resistance. Among these 130 cases, those with disputed rpoB mutation on rpoB sequencing analysis were enrolled in the present study. The medical records of these patients with M. tuberculosis isolates with disputed rpoB mutations were reviewed retrospectively in August 2016.
In addition, we identified the M. tuberculosis strains with available rpoB sequencing results from the International Tuberculosis Research Center (ITRC) in Masan, South Korea. Over a 5-year period from 2005 and 2010, 170 strains with rpoB mutation identified via rpoB sequencing were collected at the ITRC, through the South Korean MDR-TB cohort study12. Of these 170 strains, we identified those with disputed mutations on rpoB sequencing analysis.
The study protocol was approved by the Institutional Review Board of Asan Medical Center, which waived the requirement for informed consent due to the retrospective nature of the analysis.
The MTBDRplus assay was performed at our center according to the manufacturer's instructions13. The attending physician made the decision regarding whether to perform an MTBDRplus assay.
DNA sequencing was performed using a polymerase chain reaction (PCR)–direct sequencing system (Cosmo Co., Seoul, Korea). A 501-bp fragment of the rpoB gene was sequenced. The following forward and reverse primers (5′–3′) were used: TCAAGGAGAAGCGCTACGACCTGGC and ACGGGTGCACGTCGCGGACCTCCA. The primers were synthesized by JieLi Bio Co. (Shanghai, China). The PCR conditions included denaturation at 94℃ for 10 minutes, followed by 35 cycles of amplification at 94℃ for 30 seconds, 60℃ for 30 seconds, and 72℃ for 30 seconds. The PCR mixtures were prepared using 2× GoldStar Best MasterMix (CWBio Co., Beijing, China). The PCR products were used as templates for targeted DNA sequencing. To confirm the sequence-amplified PCR product, direct sequencing was performed using an ABI 373 DNA analyzer (Applied Biosystems, Foster City, CA, USA) according to the protocol supplied by the manufacturer, along with the BigDye Terminator v3.1 Cycling Sequencing Kit (Applied Biosystems). Sequencing data were assembled and analyzed using BioEdit software (Isis Pharmaceuticals, Inc., Carlsbad, CA, USA). The mutations were determined via comparison with the H37Rv sequence of the rpoB genes from the Gen-Bank database (http://www.ncbi.nih.gov/gene). The decision regarding whether to perform a DNA sequencing analysis was made by the attending physician.
Acid-fast bacillus (AFB) smears were examined following Ziehl-Neelsen staining. The AFB culture was carried out using both solid (Ogawa medium; Korean Institute of Tuberculosis, Korea) and liquid (BACTEC 960 Mycobacterial Growth Indicator Tube; Becton Dickinson, Sparks, MD, USA) media. Positive liquid media cultures or colonies on the solid medium were subjected to Ziehl-Neelsen staining and a PCR assay using Seeplex TB detection (Seegen, Seoul, Korea) to differentiate between the M. tuberculosis complex and non-tuberculous mycobacteria.
When the growth of M. tuberculosis was detected in the cultures, DST was requested according to the Korean national TB guidelines. DSTs were performed using the absolute concentration method on Löwenstein-Jensen medium at the Korean Institute of Tuberculosis, which is the reference laboratory for TB in South Korea. Wells contained the following critical concentrations of anti-TB drugs: 0.2 µg/mL of INH and 40 µg/mL of RIF. Growth exceeding that of the control wells by >1% was considered to indicate drug resistance.
Patient treatment was not standardized. Treatment was individualized according to the previous treatment history and DST results for other drugs, such as INH. The treatment outcome categories were assessed in accordance with the revised 2013 WHO definitions14. In addition, we assessed the incidence of recurrence within 1 year following the completion of treatment.
Among the 130 cases with RIF resistance on the MTBDRplus assay, 15 were found to be RIF-susceptible on DSTs. Of these 15 cases, 11 underwent rpoB sequencing analysis. With the exception of two cases with wild-type strains on rpoB sequencing, the remaining nine exhibited disputed rpoB mutations (Figure 1). Thus, disputed rpoB mutations were identified in at least 6.9% of the cases (9/130) with RIF resistance, as determined via the MTBDRplus assay, at our center. In addition, the sequencing of the rpoB gene of 170 strains with rpoB mutations at the ITRC showed that 23 (13.5%) had disputed rpoB mutations12.
Table 1 presents the results of the MTBDRplus assay and rpoB sequencing for the nine patients with rpoB mutations in their M. tuberculosis isolates. Among these nine patients, the most common disputed mutation was CTG511CCG (n=5), followed by GAC516TAC (n=2) and CAC526AAC (n=2). These sequencing patterns were precisely consistent with the location of the wild-type loss indicated in the MTBDRplus assay. In addition, the order of frequency of the disputed rpoB mutations in the 23 strains stored in the ITRC was as follows: CTG533CCG (n=8), GAC516TAC (n=6), CAC526CTC (n=4), CTG511CCG (n=3), CAC526AAC (n=1), and ATG515GTG (n=1).
After combining the results of the 32 strains from both our center and the ITRC, the order of frequency of the disputed rpoB mutations was as follows: CTG511CCG (n=8), GAC516TAC (n=8), CTG533CCG (n=8), CAC526CTC (n=4), CAC526AAC (n=3), and ATG515GTG (n=1).
Table 2 summarizes the treatment regimen, duration, and treatment outcome of the nine patients with disputed rpoB mutations at our center. All of these patients were diagnosed with pulmonary TB and more than 50% were treated with an MDR-TB regimen (patients 1, 3, 6, 7, 8, and 9). The remaining three patients received a treatment regimen comprising first-line drugs (patients 2, 4, and 5). In particular, two patients (patients 4 and 5) were prescribed a regimen including high-dose RIF (20 mg/kg). Most of the patients (except patient 7) exhibited favorable outcomes. We could not assess the treatment outcomes of 23 cases with disputed mutations in the ITRC.
Among rpoB mutations, the most commonly detected have been GAC516GTC, CAC526TAC, CAC526GAC, and TCG531TTG, all of which are included as mutation probes in the MTBDRplus assay. In contrast to these four undisputed mutations that cause high-level RIF resistance, some rpoB mutations can cause low-level RIF resistance and are referred to as disputed mutations7. The most important finding of this study was that disputed rpoB mutations do not appear to be rare among those exhibiting RIF resistance in South Korea.
Van Deun et al.15 reported that disputed mutations comprise 13.1% and 10.6% of all rpoB mutations in strains derived from patients with first failure or relapse in Bangladesh and Kinshasa, respectively. In a recent study, most of the non-silent RRDR mutations detected directly from diagnostic sputum specimens of previously untreated cases in Bangladesh belonged to the disputed group7. Another study conducted in Australia reported that M. tuberculosis isolates that are genotypically resistant but phenotypically susceptible to RIF accounted for 9% of all cases5. A recent study performed in Germany reported that 2.8% of strains (4/143) submitted to the German National Reference Laboratory had disputed rpoB mutations in INH-resistant and RIF-susceptible M. tuberculosis isolates16. In the present study, a disputed rpoB mutation was identified as the cause of phenotypic or genotypic RIF resistance from at least 6.9% (our center) and 13.5% (ITRC) cases. It should be noted that we could not assess the exact prevalence of the disputed rpoB mutations at our center, because rpoB sequencing was not performed for all cases, as it was dependent on the attending physician.
Not all disputed mutations exhibit low-level resistance. One previous study reported that most strains with disputed mutations were RIF-resistant in a routine Löwenstein-Jensen medium DST (78.7%)15. Given that the strains with CTG511CCG or CTG533CCG were found to be RIF-susceptible by a phenotypic DST in approximately half of the tested cases15171819, the level of resistance appears to be dependent on the type of the disputed mutation. As rpoB sequencing was conducted for only a portion of the cases that were RIF-resistant by the MTBDRplus assay at our center, it is highly likely that we overlooked the disputed rpoB mutations with high-level resistance, which would have been reported as RIF-resistant in the DST. In fact, among the 115 strains that were RIF-resistant both by the MTBDRplus assay and DST, 17 exhibited a pattern suspected as being a disputed mutation on the MTBDRplus assay (the loss of the wild-type probe without any hybridization of the mutation probe). Therefore, the prevalence of disputed rpoB mutations may have been at lot higher if some of these 17 cases actually had disputed rpoB mutations; as high as 20.0% (26 out of 130 stains with RIF resistance on the MTBDRplus assay).
Although the WHO guidelines recommend that any patient with RIF-resistant TB should be treated with the recommended MDR-TB regimen irrespective of the INH susceptibility20, patients infected with M. tuberculosis strains exhibiting low-level RIF resistance caused by disputed mutations present a therapeutic challenge in terms of high-dose RIF based treatment. Van Ingen et al.6 reported the feasibility of high-dose RIF therapy for the GAC516TAC mutation, with an MIC of 0.5–2 µg/mL, determined using 7H10 agar dilution methods. They found that a 20 mg/kg dose of RIF is likely to overcome an MIC of 1 µg/mL, but not 2 µg/mL, thus suggesting that high-dose RIF treatment (20 mg/kg) might be feasible for isolates with disputed mutations for an MIC up to 1 µg/mL6. In fact, evidence in the literature for the current standard dose of RIF (600 mg) is scarce, and a high-dose RIF has been shown to increase bactericidal activity without significantly increasing the toxicity21. Therefore, the use of a higher dose of RIF (e.g., 900 mg or 1,200 mg) should be re-explored in the context of low-level RIF resistance5. In our present study, two patients (patients 4 and 5) were treated with a high dose of a RIF-containing regimen, and showed favorable outcomes. Although there may be concerns regarding the potential hazardous adverse effects of higher RIF doses, there is little evidence that higher daily RIF doses lead to increased toxicity in the literature21. In fact, one randomized phase II trial recently found that there was no significant increase in adverse events when the RIF dose was increased from 10 to 15 or 20 mg/kg22. Nevertheless, further studies are needed to determine the efficacy and optimal duration of a high-dose RIF-based regimen for disputed rpoB mutations with low-level resistance.
Previous studies have shown that M. tuberculosis isolates with disputed rpoB mutations had poorer outcomes if they were treated with standard dosing RIF-based regimens; in fact, the rate of treatment failure ranged from 25% to 70%, even though the cases were found to be RIF-susceptible via DST52324. Therefore, in cases found to be RIF-resistant via an MTBDRplus assay and RIF-susceptible via the DST, rpoB sequencing should be performed to identify the cause of the discrepancy, particularly regarding the presence of disputed mutations. If rpoB sequencing cannot be performed, treatment options other than the standard first-line anti-TB regimens (e.g., regimens containing a higher dose of RIF or second-line drugs) should be considered. In addition, considering that this resistance pattern is not detected by the current phenotypic DST methods, it may be necessary to perform genotypic DST for all M. tuberculosis isolates, if the burden of disputed rpoB mutations cannot be ignored. In this context, we have developed a rapid molecular test kit, which can rapidly detect disputed mutations without the need for sequencing analysis (Supplementary Methods, Supplementary Figures S1, S2).
Our study had several notable limitations. Most significantly, it was conducted with strains from only two centers, and thus included only a few cases with disputed rpoB mutations. The number of strains from these centers appears to be insufficient to draw highly reliable conclusions regarding the frequency and common types of mutations in South Korea. In addition, as the decision for performing sequencing analysis was dependent on the attending physician, gene sequencing was not performed in some cases with discrepant results at our center. Moreover, we did not measure the MIC of each strain with disputed mutations. Finally, the isolates yielding discrepant results were not re-tested by all the methods to ascertain the reproducibility of the initial testing result.
In conclusion, disputed rpoB mutations do not appear to be rare among the M. tuberculosis strains exhibiting RIF resistance in South Korea. We have identified the types of disputed rpoB mutations from 32 strains with this mutation at our center and the ITRC. All the patients with strains showing disputed mutations received individualized treatment regimens at our center, with favorable outcomes observed in most cases, including two that were treated with a high-dose RIF-containing regimen.
Acknowledgments
This study was supported by a 2015 Grant from The Korean Academy of Tuberculosis and Respiratory Diseases.
References
1. Park JS. Issues related to the updated 2014 Korean guidelines for tuberculosis. Tuberc Respir Dis. 2016; 79:1–4.

2. Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis. 2015; 19:1276–1289. PMID: 26467578.
3. Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993; 341:647–650. PMID: 8095569.
4. Ocheretina O, Escuyer VE, Mabou MM, Royal-Mardi G, Collins S, Vilbrun SC, et al. Correlation between genotypic and phenotypic testing for resistance to rifampin in Mycobacterium tuberculosis clinical isolates in Haiti: investigation of cases with discrepant susceptibility results. PLoS One. 2014; 9:e90569. PMID: 24599230.
5. Ho J, Jelfs P, Sintchencko V. Phenotypically occult multidrug-resistant Mycobacterium tuberculosis: dilemmas in diagnosis and treatment. J Antimicrob Chemother. 2013; 68:2915–2920. PMID: 23838950.
6. van Ingen J, Aarnoutse R, de Vries G, Boeree MJ, van Soolingen D. Low-level rifampicin-resistant Mycobacterium tuberculosis strains raise a new therapeutic challenge. Int J Tuberc Lung Dis. 2011; 15:990–992. PMID: 21682979.
7. Van Deun A, Aung KJ, Hossain A, de Rijk P, Gumusboga M, Rigouts L, et al. Disputed rpoB mutations can frequently cause important rifampicin resistance among new tuberculosis patients. Int J Tuberc Lung Dis. 2015; 19:185–190. PMID: 25574917.
8. Huitric E, Werngren J, Jureen P, Hoffner S. Resistance levels and rpoB gene mutations among in vitro-selected rifampin-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother. 2006; 50:2860–2862. PMID: 16870787.
9. Cavusoglu C, Turhan A, Akinci P, Soyler I. Evaluation of the genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis isolates. J Clin Microbiol. 2006; 44:2338–2342. PMID: 16825346.
10. Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998; 79:3–29. PMID: 10645439.
12. Song T, Park Y, Shamputa IC, Seo S, Lee SY, Jeon HS, et al. Fitness costs of rifampicin resistance in Mycobacterium tuberculosis are amplified under conditions of nutrient starvation and compensated by mutation in the beta' subunit of RNA polymerase. Mol Microbiol. 2014; 91:1106–1119. PMID: 24417450.
13. Hain Lifescience. Genotype MTBDRplus ver 2.0 [Internet]. Nehren: Hain Lifescience;2017. cited 2017 Jan 2. Available from: http://www.hain-lifescience.de/en/products/microbiology/mycobacteria/tuberculosis/genotype-mtbdrplus.html.
14. Definitions and reporting framework for tuberculosis: 2013 revision [Internet]. Geneva: World Health Organization;2017. cited 2017 Jan 2. Available from: http://apps.who.int/iris/bitstream/10665/79199/1/9789241505345_eng.pdf.
15. Van Deun A, Aung KJ, Bola V, Lebeke R, Hossain MA, de Rijk WB, et al. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol. 2013; 51:2633–2640. PMID: 23761144.

16. Andres S, Hillemann D, Rusch-Gerdes S, Richter E. Occurrence of rpoB mutations in isoniazid-resistant but rifampin-susceptible Mycobacterium tuberculosis isolates from Germany. Antimicrob Agents Chemother. 2014; 58:590–592. PMID: 24145520.
17. Kambli P, Ajbani K, Sadani M, Nikam C, Shetty A, Udwadia Z, et al. Defining multidrug-resistant tuberculosis: correlating GenoType MTBDRplus assay results with minimum inhibitory concentrations. Diagn Microbiol Infect Dis. 2015; 82:49–53. PMID: 25749461.
18. Williams DL, Spring L, Collins L, Miller LP, Heifets LB, Gangadharam PR, et al. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998; 42:1853–1857. PMID: 9661035.
19. Yakrus MA, Driscoll J, Lentz AJ, Sikes D, Hartline D, Metchock B, et al. Concordance between molecular and phenotypic testing of Mycobacterium tuberculosis complex isolates for resistance to rifampin and isoniazid in the United States. J Clin Microbiol. 2014; 52:1932–1937. PMID: 24648563.
20. WHO treatment guidelines for drug-resistant tuberculosis, 2016 update [Internet]. Geneva: World Health Organization;2016. cited 2017 Jan 2. Available from: http://www.who.int/tb/MDRTBguidelines2016.pdf.
21. van Ingen J, Aarnoutse RE, Donald PR, Diacon AH, Dawson R, Plemper van Balen G, et al. Why do we use 600 mg of rifampicin in tuberculosis treatment? Clin Infect Dis. 2011; 52:e194–e199. PMID: 21467012.

22. Jindani A, Borgulya G, de Patino IW, Gonzales T, de Fernandes RA, Shrestha B, et al. A randomised Phase II trial to evaluate the toxicity of high-dose rifampicin to treat pulmonary tuberculosis. Int J Tuberc Lung Dis. 2016; 20:832–838. PMID: 27155189.

23. Williamson DA, Roberts SA, Bower JE, Vaughan R, Newton S, Lowe O, et al. Clinical failures associated with rpoB mutations in phenotypically occult multidrug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2012; 16:216–220. PMID: 22137551.
24. Pang Y, Ruan YZ, Zhao J, Chen C, Xu CH, Su W, et al. Diagnostic dilemma: treatment outcomes of tuberculosis patients with inconsistent rifampicin susceptibility. Int J Tuberc Lung Dis. 2014; 18:357–362. PMID: 24670576.

Supplementary Material
Supplementary material can be found in the journal homepage (http://www.e-trd.org).
Supplementary Figure S1
(A) A schematic illustration of each probe from the upgraded version of the MolecuTech REBA MTB-MDR Kit. (B) Detection of the CTG511CCG (L511P) disputed rpoB mutation using the upgraded version of the MolecuTech REBA MTB-MDR Kit.
Supplementary Figure S2
The detection of undisputed (A, B) and disputed (C–F) mutations using the upgraded version of the MolecuTech REBA MTB-MDR Kit.
Figure 1
Study flow chart. DST: drug susceptibility test; NTM: non-tuberculous mycobacteria; AFB: acid-fast bacillus; RIF: rifampin.
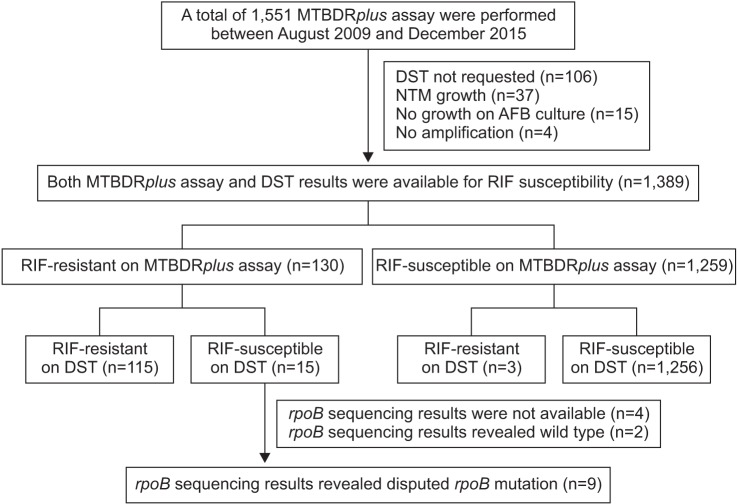
Table 1
Results of the MTBDRplus assay and rpoB sequencing in the nine study patients with disputed rpoB mutations
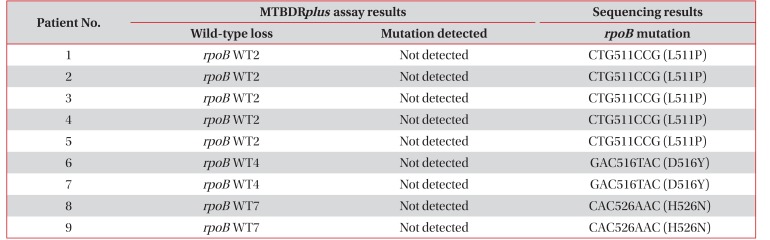
Table 2
Isoniazid susceptibility, treatment regimen and duration, and treatment outcome in the nine study patients with disputed rpoB mutations
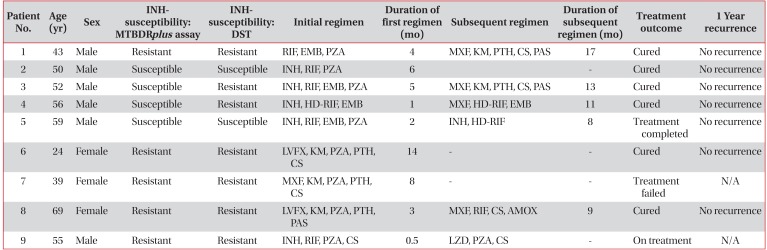
INH: isoniazid; DST: drug susceptibility test; RIF: rifampicin; EMB: ethambutol; PZA: pyrazinamide; MXF: moxifloxacin; KM: kanamycin; PTH: prothionamide; CS: cycloserine; PAS: p-aminoslicylic acid; HD-RIF: high dose rifampin; LVFX: levofloxacin; N/A: not available; AMOX: amoxicillin/clavulanate; LZD: linezolid.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download