Abstract
Background
Airway epithelial cells are the first line of defense, against pathogens and environmental pollutants, in the lungs. Cellular stress by cadmium (Cd), resulting in airway inflammation, is assumed to be directly involved in tissue injury, linked to the development of lung cancer, and chronic obstructive pulmonary disease (COPD). We had earlier shown that ACN9 (chromosome 7q21), is a potential candidate gene for COPD, and identified significant interaction with smoking, based on genetic studies. However, the role of ACN9 in the inflammatory response, in the airway cells, has not yet been reported.
Methods
We first checked the anatomical distribution of ACN9 in lung tissues, using mRNA in situ hybridization, and immunohistochemistry. Gene expression profiling in bronchial epithelial cells (BEAS-2B), was performed, after silencing ACN9. We further tested the roles of ACN9, in the intracellular mechanism, leading to Cd-induced production, of proinflammatory cytokines in BEAS-2B.
Results
ACN9 was localized in lymphoid, and epithelial cells, of human lung tissues. ACN9 silencing, led to differential expression of 216 genes. Pathways of sensory perception to chemical stimuli, and cell surface receptor-linked signal transduction, were significantly enriched. ACN9 silencing, further increased the expression of proinflammatory cytokines, in BEAS-2B after Cd exposure.
The prevalence of chronic obstructive pulmonary disease (COPD) is increasing, and it is expected to become the fourth cause of death worldwide by 20301. To date, the molecular mechanisms leading to the development of COPD are poorly understood, and there are currently no effective therapies that can prevent the pathogenesis of COPD. Therefore, finding new biomarkers and understanding the biological mechanism of COPD are essential to reduce the disease morbidity and mortality.
COPD is characterized by airflow limitation that is not fully reversible and an abnormal inflammatory response in the lung. Previous studies demonstrated that environmental risk factors, including cigarette smoking, air pollution, and chemicals, contributed to the development of COPD234. Although cadmium (Cd) is a constituent of cigarette smoke, the precise role of Cd in COPD is not fully defined. However, growing evidence indicates that Cd may play a key role in smoking-induced disorders, including impaired lung function, chronic inflammation, or the development of emphysema5678.
In our previous genetic association study on patients with COPD, we identified candidate disease susceptibility loci. Notably, a number of single nucleotide polymorphisms were identified in the ACN9 gene, of which the risk alleles of rs10231916 and rs10229181 were associated with attenuated expression of ACN9 in the lung9. These findings suggest that altered expression of ACN9 may contribute to human lung pathological conditions. Nevertheless, the biological function or the pathogenic roles of ACN9 have not been widely studied. The human ACN9 gene (sdh7 in yeast, SDHAF3 in humans) encompasses a 65,174 bp region at chromosome 7q22.1 (chr 7:97,116,593–97,181,763) and consists of two exons with protein-coding exons. It belongs to the LYR motif protein family, which is localized in the mitochondrial matrix and is related to Fe-S cluster protection in succinate dehydrogenase (SDH) maturation in yeast and Drosophila1011.
In this study, we determined the functional roles of ACN9 by mRNA profiling after knocking down ACN9 expression. We also investigated whether ACN9 has a role in the inflammatory signaling pathway in airway epithelial cells.
Human lung tissue samples were provided from Biobank of the Kangwon National University Hospital (KNUH) in accordance with an approved protocol by Institutional Review Board of KNUH (2014-12-014). Human lungs and other tissues from the surgical specimens were fixed in 10% formalin solution at 60℃ overnight and embedded in paraffin. After deparaffinization with graded concentrations of xylene and ethanol, slides were immersed in 3% H2O2 in phosphate-buffered saline (PBS; Thermo Fisher Scientific Inc., Waltham, MA, USA) for 30 minutes at room temperature to block endogenous peroxidase activity. Then, they were incubated with 3% bovine serum albumin (BSA) for 2 hours to reduce non-specific background staining. The sections were then incubated with monoclonal antibody against ACN9 (1:50, Abcam, Cambridge, UK) in PBS with BSA.
In situ hybridization was performed using formalin-fixed paraffin-embedded (FFPE) tissues using the RNAScope 2.0 RED assay, according to the manufacturer's instructions (#310036; Advanced Cell Diagnostics Inc., Hayward, CA, USA). First, 4-µm sections were cut from the FFPE blocks and mounted on Superfrost Plus microscope slides (Thermo Fisher Scientific Inc.). Slides were placed at 60℃ for 1 hour in a dry oven. Mounted sections were deparaffinized in xylene, dehydrated in 100% ethanol, and then treated serially with ACD pretreatment 1 (endogenous hydrogen peroxidase block) for 10 minutes at room temperature, ACD pretreatment 2 (boiling in citrate buffer) for 8–12 minutes at 98℃–100℃, and ACD pretreatment 3 (protease digestion) for 12–30 minutes at 40℃. Pretreatment 2 and 3 conditions for each paraffin block were optimized to maximize the signal-to noise ratio using internal control (Hs-PPIB, #313901; Advanced Cell Diagnostics Inc.) and negative control (Fast Red, #310043; Advanced Cell Diagnostics Inc.) RNA probes. Next, hybridization between the target RNA and the selected ACD probe (Hs-SDHAF3, #441611; Advanced Cell Diagnostics Inc.) was performed during a 2-hour incubation period.
Unless otherwise indicated, all materials for cell culture were purchased from Gibco (Thermo Fisher Scientific Inc.). The human bronchial epithelial cell line (BEAS-2B) was kindly provided by Biomedical Research Institute at Seoul National University Hospital. BEAS-2B cells were maintained in defined keratinocyte serum-free medium containing epidermal growth factor, 100 U/mL of penicillin, and 100 µg/mL of streptomycin. Cell cultures were incubated at 37℃ in humidified atmosphere containing 5% CO2. BEAS-2B cells were transfected with human ACN9 siRNA (ACN9 ON-TARGET plus SMART pool) and/or negative control siRNA (ON-TARGET plus non-targeting pool) at a final concentration of 15 nM in the presence of DharmaFECT reagent (Dharmacon, Lafayette, CO, USA), as per the manufacturer's protocol. After transfection, the cells were incubated with or without 10 µM Cd for 24 hours for quantitative polymerase chain reaction analysis and Western blotting5. Cd was dissolved in sterile water and stored as a stock solution. It was diluted in cell media to reach the indicated concentration.
BEAS-2B cells (2×106 cells/well) in 6-well culture plates were treated in the absence or presence of Cd (Sigma, St. Louis, MO, USA) for 24 hours. Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed to first-stand complementary DNA (cDNA) using random primer (9-mer) and QuantiTect reverse transcriptase (Qiagen, Hilden, Germany), according to the manufacturer's protocols. Transcripts were quantitated using Power SYBR Green PCR (Applied Biosystems, Foster City, CA, USA) and the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). Quantitation was normalized with 18s rRNA (internal control). Quantitative real time polymerase chain reaction was performed using primer sequences listed in Table 1. For quality control, RNA purity and integrity were evaluated by OD 260/280 ratio and analyzed by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
To quantitate the secreted interleukin 6 (IL-6) levels, the cell culture supernatants of BEAS-2B cells were measured by human IL-6 Quantikine ELISA Kit (R&D Systems, Minneapolis, MN, USA). The enzyme-linked immunosorbent assay plates were read using a microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Cell lysates were separated on an sodium dodecyl sulfate polyacrylamide gel electrophoresis gel (10% or 15%), transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA), and blocked with 3% skim milk. The membrane was probed with primary antibodies against antihuman interleukin 1β (IL-1β) antibody (AF-201-NA; R&D Systems), anti–IL-1 RA antibody (SC-25444; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti–mitogen-activated protein kinase (MAPK) signal pathway sampler kit antibody (#9913; Cell Signaling Technologies, Danvers, MA, USA), and anti–β-actin antibody (SC-47778, Santa Cruz Biotechnology) overnight at 4℃. The membranes were further probed with horseradish peroxidase–conjugated secondary anti-sera (A9917, A6667, or A5420; Sigma) and visualized by Power-Opti ECL solution (Bionote, Hwaseong, Korea) and a cooled CCD camera system (Bio-Rad Laboratories Inc., Hercules, CA, USA).
The Affymetrix Whole transcript Expression Array process was executed according to the manufacturer's protocol (GeneChip Whole Transcript PLUS Reagent Kit; Affymetrix, Santa Clara, CA, USA). cDNA was synthesized using the GeneChip WT (whole transcript) Amplification Kit (Affymetrix) as described by the manufacturer. The sense cDNA was then fragmented and biotin-labeled with terminal deoxynucleotidyl transferase (TdT) using the GeneChip WT Terminal Labeling Kit (Affymetrix). Approximately 5.5 µg of labeled DNA target was hybridized to the Affymetrix GeneChip Human 2.0 ST Array at 45℃ for 16 hours. Hybridized arrays were washed and stained on a GeneChip Fluidics Station 450 and scanned on a GCS3000 Scanner (Affymetrix). Signal values were computed using the Affymetrix GeneChip Command Console software.
Raw data were extracted automatically in Affymetrix data extraction protocol using the software provided by Affymetrix GeneChip Command Console software. After importing CEL files, the data were summarized and normalized with robust multi-average (RMA) method implemented in Affymetrix Expression Console software. We exported the result with gene-level RMA analysis and performed the differentially expressed gene (DEG) analysis. Statistical significance of the expression data was determined using fold change. For a DEG set, hierarchical cluster analysis was performed using complete linkage and Euclidean distance as a measure of similarity. Gene-enrichment and functional annotation analysis for significant probe list was performed using DAVID (http://david.abcc.ncifcrf.gov/home.jsp). All data analyses and visualization of DEGs were conducted using R 3.1.2 (http://www.r-project.org). Statistical analyses were performed with t test for multiple groups using GraphPad Prism (GraphPad Software, San Diego, CA, USA). p-value is indicated in the figure.
We investigated cellular localization of ACN9 expression in human tissues using both in situ hybridization and immunohistochemistry. We observed positive staining for ACN9 within the bronchial epithelium and lymphoid-like cells in normal lung tissues (Figure 1A). We next assessed the protein expression of ACN9 in human tissues, including the liver and lung tissues (Figure 1B). Our expression patterns revealed a significant expression of ACN9 in human lung tissues.
To identify endogenous ACN9 target genes, we transiently transfected siACN9 into bronchial epithelial cells. To test the efficiency of the ACN9 transcript suppression, BEAS-2B cells were transfected with 15 nM siRNA and cultured for 24 hours. Then, the cells were processed for Western blot analysis to determine the expression of ACN9. As expected, transfection of BEAS-2B cells with 15 nM siRNA resulted in approximately 70% silencing of ACN9 gene expression levels compared with control (Figure 2A). Cd treatment further decreased ACN9 expression (Figure 2B). Microarray results identified genes that were significantly altered by ACN9 knock down with or without Cd (>2.0-fold, p<0.05) (Figure 2C). We identified 113 upregulated genes and 103 downregulated genes as a result of ACN9 knockdown in BEAS-2B. We also identified 408 up-regulated genes and 251 downregulated genes in response to Cd treatment after ACN9 silencing (Figure 2D). We found that the GO categories of “defense response,” “inflammatory response,” and “innate immune response” were significantly enriched with Cd treatment, while “cell surface receptor” and “sensory perception to chemical” were ranked at the top list in ACN9 silencing (Figure 2E).
To investigate the role of ACN9 in the inflammatory responses of human bronchial epithelial cells, BESA-2B cells were treated with Cd after transfection with siACN9. Silencing ACN9 before the addition of Cd enhanced the inflammatory responses compared to that in Cd-treated control cells (Figure 3A, B). These results showed that silencing ACN9 significantly promoted the Cd-induced proinflammatory cytokine production in BEAS-2B cells. We next determined whether ACN9 silencing has an effect on Cd-induced MAPK activity. After silencing ACN9, JNK, and p38/MAPK pathway was further activated by Cd treatment in BEAS-2B cells, while ERK activation was not influenced by ACN9 silencing (Figure 3C, D).
In this study, ACN9 gene and proteins were found in normal lung epithelial cells and lymphoid-like cells. Gene expression profiling revealed that sensory perception to chemical pathway was altered after ACN9 knockdown in bronchial epithelial cells. Expression of inflammatory cytokines further increased following exposure to Cd in bronchial epithelial cells with ACN9 gene knockdown. We also demonstrated that this may be associated with the p-38/MAPK pathway. Based on the above evidences, ACN9 might be a candidate gene in COPD because ACN9 silencing in cells increased the inflammatory pathways by Cd.
ACN9 belongs to the LYR motif protein family that support the assembly of SDH under normal physiological conditions. Previous studies have shown that the functions of several other LYR motif proteins are linked to diseases such as insulin resistance, muscular hypotonia, deficiency of multiple mitochondrial oxidative phosphorylation complex, and alcohol dependence12131415. ACN9 is a central component in the ancient Fe-S cluster metabolism and in protecting SDHB from oxidative stress during the assembly process in Drosophila16. High concentrations of reactive oxygen species (ROS) damage Fe-S cluster, and protection of the sensitive Fe-S clusters is important in the presence of oxidative stress. Respiratory chain complexes I, II, and III play a role in producing a significant amount of ROS in mitochondria. Deletion of ACN9 caused a dramatic SDH deficiency with muscular and neuronal defects that are suggestive of neurodegeneration observed in humans with mutations in SDHAF1101117. Until now, only a few studies have investigated the effects of ACN9 on inflammatory response and mitochondrial dysfunction18, and no studies have been performed in human lung epithelial cells. In the present study, we used in situ hybridization and immunohistochemistry to investigate the cellular localization of ACN9 mRNA and protein in human lung tissues. Our results indicate that ACN9 expression was detected in both airway epithelium and lymphoid cells in the lung tissues.
The airway epithelium is the barrier between inhaled air containing toxic compounds, including Cd, and the underlying lung tissue. To maintain this barrier, continuous cell replacement and repair of the epithelium are of crucial importance, while noxious agents are most important risk factor for airway diseases. In general, Cd exposure causes accumulation in humans at low concentrations over long period. Therefore, we selected 10 µM of Cd dose in bronchial epithelial cells as in vitro model for airway exposure to the noxious agent. The final consequence of the Cd-induced electron transport chain blockade is the loss of capacity of the mitochondria to generate ATP. Exposure to Cd results in the increased formation of reducing equivalents (NADH) by the citric acid cycle and increased mitochondrial oxygen consumption and ATP formation via oxidative phosphorylation19. This could be due to the direct action of Cd on mitochondrial function, which mediates the inhibition of electron transfer, disruption of the respiratory complexes, and membrane permeability. Recent studies have suggested that an abnormality in mitochondrial function is associated with development of lung diseases202122. The large amount of free radicals present in inhaled Cd will initially be able to induce an apoptotic signal in the exposed cells23.
We investigated the genomic response to Cd exposure in bronchial epithelial cells after ACN9 silencing using microarray to understand the molecular mechanism of ACN924. In our study, several genes were notably upregulated or down-regulated in ACN9 knockdown cells with or without Cd stress as compared to that in control cells. Cd stress was associated with inflammatory responses, while ACN9 silencing in bronchial epithelial cells was associated with pathways such as sensory perception to chemical and cell surface receptor-linked signal transduction. We further tested the roles of ACN9 in the intracellular mechanism of exposure to Cd in terms of proinflammatory cytokines in bronchial epithelial cells (BEAS-2B). We observed that ACN9 silencing significantly increased the expression of proinflammatory cytokines at mRNA and translational levels when BEAS-2B cells were treated with Cd after ACN9 silencing. Furthermore, ACN9 silencing led to further activation of the p38/MAPK pathway induced by Cd-treated cells.
Because ACN9 is related to SDH, a possible mechanism of the inflammatory response induced by ACN9 silencing is via SDH that is involved in the inflammatory response25. Previous reports have shown that SDH mutations display elevated levels of succinate in immune cells. Succinate stabilizes hypoxiainducible factor-1α (HIF-1α) by inhibiting prolyl hydroxylase domain enzyme activity. Stabilized HIF1-α binds to HIF response elements in target genes, including those encoding glycolytic enzymes, angiogenic factors, and the inflammatory cytokine IL-1β, thereby exacerbating inflammation26. We speculated that ACN9 silencing diminished the consumption of oxygen and production of ATP. High succinate concentrations have been detected in the plasma of patients. In addition, patients harboring mutations in SDH exhibited increased HIF-1α activity272829. Further study is needed to investigate the mechanisms by which succinate enhances proinflammatory cytokine production in BEAS-2B cells.
There are some limitations related to our study. First, we measured several inflammatory cytokines at gene levels by real-time polymerase chain reaction. Although we measured IL-6 in the supernatant, other protein-level inflammatory reactions were not measured. Second, we studied bronchial cell models, but several cell types may be involved in the pathogenesis of COPD. Further in vivo model should be studied to further understand the role of ACN9 gene.
In conclusion, our study provides new insight into the function of ACN9 in the pathogenesis of COPD. Further study is needed to understand the role of ACN9 in the inflammatory response and mitochondrial dysfunction of airway diseases.
Figures and Tables
Figure 1
ACN9 localization in human lung tissues. (A) In situ hybridization showing ACN9 mRNA localized in human lung tissues. Paraffin sections were rehydrated and incubated with an anti-ACN9 or scramble probe as negative control. Results are normalized to anti-PPIB probe (internal control) used. (B) Protein-level expression of ACN9 in the liver and lung tissues was analyzed using immunohistochemistry. Data shown is representative of at least two independent experiments.

Figure 2
Heatmaps for representative gene sets dysregulated by cadmium (Cd)-treated ACN9-silenced cells. Effect of Cd and siACN9 on the expression of ACN9 mRNA level. (A) BEAS-2B cells transfected with siACN9 (15 nM) compared to the control scrambled siRNA. (B) BEAS-2B cells (2×105 cells per well for quantitative polymerase chain reaction) were treated with the indicated concentrations of Cd (10 µM) for 24 or 48 hours. For quantitative real time polymerase chain reaction (qRT-PCR), statistical evaluation was performed using Student's t test. Data shown is representative of at least three independent experiments. (C) Comparison of gene expression patterns obtained from cDNA microarray and qRT-PCR with ACN9 knockdown in Cd-treated bronchial epithelial cells. Heat map and bars indicate the magnitude of gene expression changes. (D) Numbers of differentially expressed genes in the ACN9-silenced BEAS-2B cells with/without Cd. (E) Gene ontology analysis showing Cd and ACN9 silencing responsive relationships in terms of biological processes. *p≤0.05, **p≤0.01, and ***p≤0.001 were considered significant. Mock: transfection reagent alone; siNC: small interference RNA of non-targeting control; siACN91-3: small interference RNA for ACN9 at 10 nM, 15 nM, and 25 nM.

Figure 3
ACN9-silenced cells drive inflammatory responses. BEAS-2B cells were pretreated for 24 hours with small interference RNA for ACN9 (siACN9; 15 nM) after being stimulated with cadmium (Cd) (10 µM). (A) mRNA was extracted from total cell lysates and analyzed by quantitative polymerase chain reaction for proinflammatory cytokines. (B) Supernatants were analyzed by enzyme-linked immunosorbent assay for interleukin 6 (IL-6). (C) Whole-cell lysates were analyzed by Western blotting for proinflammatory cytokine (IL-1β) and p38/mitogen-activated protein kinase (MAPK) signal pathway and normalized by β-actin. (D) The protein expression levels of phosphorylated JNK, ERK, and p38 MAPK were calculated by normalized to total ERK, and IL-1β and IL-1RA were normalized by β-actin. For enzyme-linked immunosorbent assay and quantitative real time polymerase chain reaction, statistical evaluation was conducted, using Student's t test *p≤0.05, **p≤0.01, and ***p≤0.001 were considered significant. Data shown is representative of at least three independent experiments. TNF-α: tumor necrosis factor α; COX2: cyclooxygenase 2; iNOS: inducible nitric oxide synthase; MMP9: matrix metalloproteinase 9; siNC: small interference RNA of non-targeting control.
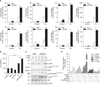
Table 1
Primer sequences used for quantitative RT-PCR
Acknowledgments
This study was supported by a 2015 Grant from the Korean Academy of Tuberculosis and Respiratory Disease. The biospecimens for this study were provided by the Kangwon National University Hospital Biobank, a member of the National Biobank of Korea, supported by the Ministry of Health and Welfare.
References
1. Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006; 3:e442.
2. Fabbri LM. Smoking, not COPD, as the disease. N Engl J Med. 2016; 374:1885–1886.
3. Peacock JL, Anderson HR, Bremner SA, Marston L, Seemungal TA, Strachan DP, et al. Outdoor air pollution and respiratory health in patients with COPD. Thorax. 2011; 66:591–596.
4. Lamprecht B, McBurnie MA, Vollmer WM, Gudmundsson G, Welte T, Nizankowska-Mogilnicka E, et al. COPD in never smokers: results from the population-based burden of obstructive lung disease study. Chest. 2011; 139:752–763.
5. Lag M, Rodionov D, Ovrevik J, Bakke O, Schwarze PE, Refsnes M. Cadmium-induced inflammatory responses in cells relevant for lung toxicity: expression and release of cytokines in fibroblasts, epithelial cells and macrophages. Toxicol Lett. 2010; 193:252–260.
6. Kirschvink N, Martin N, Fievez L, Smith N, Marlin D, Gustin P. Airway inflammation in cadmium-exposed rats is associated with pulmonary oxidative stress and emphysema. Free Radic Res. 2006; 40:241–250.
7. Bertin G, Averbeck D. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review). Biochimie. 2006; 88:1549–1559.
8. Hassan F, Xu X, Nuovo G, Killilea DW, Tyrrell J, Da Tan C, et al. Accumulation of metals in GOLD4 COPD lungs is associated with decreased CFTR levels. Respir Res. 2014; 15:69.
9. Kim WJ, Lim MN, Hong Y, Silverman EK, Lee JH, Jung BH, et al. Association of lung function genes with chronic obstructive pulmonary disease. Lung. 2014; 192:473–480.
10. Dennis RA, McCammon MT. ACN9 is a novel protein of gluconeogenesis that is located in the mitochondrial intermembrane space. Eur J Biochem. 1999; 261:236–243.
11. Na U, Yu W, Cox J, Bricker DK, Brockmann K, Rutter J, et al. The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab. 2014; 20:253–266.
12. Zhu GZ, Zhang M, Kou CZ, Ni YH, Ji CB, Cao XG, et al. Effects of Lyrm1 knockdown on mitochondrial function in 3 T3-L1 murine adipocytes. J Bioenerg Biomembr. 2012; 44:225–232.
13. Haack TB, Madignier F, Herzer M, Lamantea E, Danhauser K, Invernizzi F, et al. Mutation screening of 75 candidate genes in 152 complex I deficiency cases identifies pathogenic variants in 16 genes including NDUFB9. J Med Genet. 2012; 49:83–89.
14. Lim SC, Friemel M, Marum JE, Tucker EJ, Bruno DL, Riley LG, et al. Mutations in LYRM4, encoding iron-sulfur cluster biogenesis factor ISD11, cause deficiency of multiple respiratory chain complexes. Hum Mol Genet. 2013; 22:4460–4473.
15. Dick DM, Aliev F, Wang JC, Saccone S, Hinrichs A, Bertelsen S, et al. A Systematic single nucleotide polymorphism screen to fine-map alcohol dependence genes on chromosome 7 identifies association with a novel susceptibility gene ACN9. Biol Psychiatry. 2008; 63:1047–1053.
16. Cloonan SM, Choi AM. Mitochondria: commanders of innate immunity and disease? Curr Opin Immunol. 2012; 24:32–40.
17. Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmuller H, et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat Genet. 2009; 41:654–656.
18. Hopps E, Noto D, Caimi G, Averna MR. A novel component of the metabolic syndrome: the oxidative stress. Nutr Metab Cardiovasc Dis. 2010; 20:72–77.
19. Wang Y, Fang J, Leonard SS, Rao KM. Cadmium inhibits the electron transfer chain and induces reactive oxygen species. Free Radic Biol Med. 2004; 36:1434–1443.
20. Heinzmann A, Thoma C, Dietrich H, Deichmann KA. Identification of common polymorphisms in the mitochondrial genome. Allergy. 2003; 58:830–831.
21. Kim SR, Kim DI, Kim SH, Lee H, Lee KS, Cho SH, et al. NLRP3 inflammasome activation by mitochondrial ROS in bronchial epithelial cells is required for allergic inflammation. Cell Death Dis. 2014; 5:e1498.
22. Kang MJ, Shadel GS. A mitochondrial perspective of chronic obstructive pulmonary disease pathogenesis. Tuberc Respir Dis. 2016; 79:207–213.
23. Bowler RP, Barnes PJ, Crapo JD. The role of oxidative stress in chronic obstructive pulmonary disease. COPD. 2004; 1:255–277.
24. Koedrith P, Kim HL, Seo YR. Integrative toxicogenomics-based approach to risk assessment of heavy metal mixtures/complexes: strategies and challenges. Mol Cell Toxicol. 2015; 11:265–276.
25. Mills E, O'Neill LA. Succinate: a metabolic signal in inflammation. Trends Cell Biol. 2014; 24:313–320.
26. Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013; 496:238–242.
27. Gimenez-Roqueplo AP, Favier J, Rustin P, Rieubland C, Kerlan V, Plouin PF, et al. Functional consequences of a SDHB gene mutation in an apparently sporadic pheochromocytoma. J Clin Endocrinol Metab. 2002; 87:4771–4774.
28. Dahia PL, Ross KN, Wright ME, Hayashida CY, Santagata S, Barontini M, et al. A HIF1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005; 1:72–80.
29. Hobert JA, Mester JL, Moline J, Eng C. Elevated plasma succinate in PTEN, SDHB, and SDHD mutation-positive individuals. Genet Med. 2012; 14:616–619.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download