|
Screening and diagnosis |
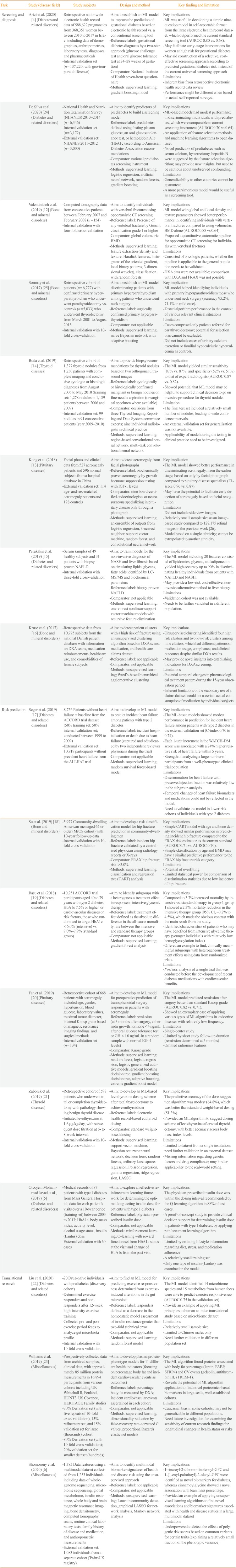
Artzi et al. (2020) [4] (Diabetes and related disorders) |
- Retrospective nationwide electronic health record data of 588,622 pregnancies from 368,351 women between 2010 to 2017 in Israel including data of demographics, anthropometrics, laboratory tests, diagnoses, and pharmaceuticals |
- Aim: to establish an ML model to improve the prediction of gestational diabetes based on electronic health record vs. a conventional screening tool |
Key implications |
|
- Internal validation set (n=137,220; with geo-temporal difference) |
- Reference labels: gestational diabetes diagnosis by a two-step approach (glucose challenge test and oral glucose tolerance test at 24–28 weeks of gestation) |
- ML was useful in developing a simple nine-question model in self-reportable format from the large electronic health record dataset, which outperformed the current standard screening tool (AUROC 0.80 vs. 0.68). |
|
- Comparator: National Institute of health seven-item questionnaire |
- May facilitate early-stage interventions for women at high risk for gestational diabetes |
|
- Methods: supervised learning; gradient boosting model |
- May aid construction of a selective, cost-effective screening approach according to predicted gestational diabetes risk instead of the current universal screening approach |
|
|
Limitations |
|
|
- Inherent bias from Retrospective electronic health record data review |
|
|
- Performance might be different when based on actual self-reported surveys. |
|
De Silva et al. (2020) [24] (Diabetes and related disorders) |
- National Health and Nutrition Examination Survey (NHANES) 2013–2014 (n=6,346) |
- Aim: to identify predictors of prediabetes to build a screening model |
Key implications |
|
- Internal validation set (n=3,172) |
- Reference label: prediabetes defined using fasting plasma glucose, an oral glucose tolerance test, or hemoglobin A1c (HbA1c) according to American diabetes Association recommendations |
- ML-based models had modest performance in discriminating inidividuals with prediabetes, which were comparable to current screening instrument (AUROC 0.70 vs 0.64). |
|
- External validation set: NHANES 2011–2012 (n=3,000) |
- Comparator: National prediabetes screening instrument |
- An application of feature selection Methods and machine learning algorithms to open dataset |
|
- Methods: supervised learning; logistic regression, artificial neural network, random forests, gradient boosting |
- Novel predictors of prediabetes such as serum calcium, hysterectomy, hepatitis B were suggested by the feature selection algorithm; may provide new insights, but need to be cautious about unobserved confounding. |
|
|
Limitations |
|
|
- Generalizability to other countries cannot be guaranteed. |
|
|
- A more parsimonious model would be useful as a screening tool. |
|
Valentinitsch et al. (2019) [12] (Bone and mineral disorders) |
- Computed tomography data from consecutive patients between February 2007 and February 2008 (n=154) |
- Aim: to identify individuals with vertebral fractures using opportunistic CT screening |
Key implications |
|
- Internal validation with four-fold cross-validation |
- Reference label: Presence of any vertebral fracture by Genant classification grade 1 or higher |
- ML model with global and local density and texture parameters showed better performance in identifying individuals with vertebral fractures compared to using volumetric BMD alone (AUROC 0.88 vs 0.64). |
|
- Comparator: global volumetric BMD |
- Proposed a quantitative, automatic pipeline for opportunistic CT screening for individuals with vertebral fractures |
|
- Methods: supervised learning; feature extraction (density and texture; Haralick features, histograms of the oriented gradient, local binary patterns, 3-dimensional wavelet), classification with random forests |
Limitations |
|
|
- Consisted of oncologic patients; whether the pipeline is applicable to the general population needs to be validated. |
|
|
- DXA data were not available; comparison with DXA and FRAX was not possible. |
|
Somnay et al. (2017) [25] (Bone and mineral disorders) |
- Retrospective cohort of patients (n=6,777) with confirmed primary hyperparathyroidism who underwent parathyroidectomy vs. controls (n=5,033) who underwent thyroidectomy from March 2001 to August 2013 |
- Aim: to establish an ML model discriminating patients with primary hyperparathyroidism among patients who underwent neck surgery |
Key implication |
|
- Internal validation with 10-fold cross-validation |
- Reference label: surgically confirmed primary hyperparathyroidism |
- ML model helped identifying individuals with primary hyperparathyroidism those who underwent neck surgery (accuracy 95.2%; 71.1% in mild case). |
|
- Comparator: not applicable |
- Tested algorithm performance in the context of various relevant clinical situations |
|
- Methods: supervised learning; naive Bayesian network with adaptive boosting |
Limitation |
|
|
- Cases comprised only patients referred for parathyroidectomy; potential for selection bias cannot be excluded. |
|
|
- Did not include cases of urinary calcium excretion or familial hypocalciuric hypercalcemia as controls. |
|
Buda et al. (2019) [14] (Thyroid diseases) |
- Retrospective cohort of 1,377 thyroid nodules from 1,230 patients with complete imaging and conclusive cytologic or histologic diagnoses from August 2006 to May 2010 (training set: 1,278 nodules in 1,139 patients between 2006 and 2009) |
- Aim: to provide biopsy recommendations for thyroid nodules based on two orthogonal ultrasound images |
Key implication |
|
- Internal validation set: 99 nodules in 91 consecutive patients (year 2009–2010) |
- Reference label: cytologically or histologically confirmed malignant or benign nodules on fine-needle aspiration (or surgical specimen where available) |
- The ML model yielded similar sensitivity (87% vs. 87%) and specificity (52% vs. 51%) to that of expert radiologists (AUROC 0.87 vs. 0.82). |
|
- Comparator: decisions from three Thyroid Imaging Reporting and Data System committee experts; nine individual radiologists in clinical practice |
- Showed potential that ML model may be helpful to support clinical decision to go on invasive procedure for thyroid nodule. |
|
- Methods: supervised learning; region-based convolutional neural network, multi-task convolutional neural network |
Limitation |
|
|
- The final test set included a relatively small number of nodules, leading to wide confidence intervals. |
|
|
- An external validation set for generalization was not available. |
|
|
- Applicability of model during the testing in clinical practice need to be investigated. |
|
Kong et al. (2018) [13] (Pituitary diseases) |
- Facial photo and clinical data from 527 acromegaly patients and 596 normal subjects from a hospital database in China |
- Aim: to detect acromegaly from facial photographs |
Key implication |
|
- External validation set: 114 age- and sex-matched acromegaly patients and 128 controls |
- Reference label: biochemically proven acromegaly by growth hormone suppression testing with IGF-1 levels |
- The ML model showed better performance in discriminating acromegaly, from the earlier stage, based on only by facial photograph compared to pituitary disease specialists (F1-score 0.96 vs. 0.87). |
|
- Comparator: nine board-certified endocrinologists or neurosurgeons specializing in pituitary disease only through a photograph |
- May have the potential to facilitate early detection of acromegaly based on facial recognition. |
|
- Methods: supervised learning; an ensemble of outputs from logistic regression, k-nearest neighbor, support vector machine, random forest, and convolutional neural network |
Limitations |
|
|
- Did not include side view images. |
|
|
- Relatively small sample size as an image-based study compared to 128,175 retinal images in the previous work [26]. |
|
|
- Model based on a single ethnicity; cannot be extrapolated to another ethnicity. |
|
Perakakis et al. (2019) [s15] (Diabetes and related disorders) |
- Serum samples of 49 healthy subjects and 31 patients with biopsy- proven NAFLD |
- Aim: to train models for the non-invasive diagnosis of NASH and liver fibrosis based on circulating lipids, glycans, fatty acids identified by LC-MS/MS and biochemical parameters |
Key implications |
|
- Internal validation with three-fold cross-validation |
- Reference label: biopsy-proven NAFLD |
- The ML model including 20 features consisted of lipidomics, glycans, and adiponectin yielded high accuracy up to 90% in discriminating healthy individuals from patients with NAFLD and NASH. |
|
- Comparator: not applicable |
- May provide a low-risk cost-effective, non-invasive alternative method to liver biopsy. |
|
- Methods: supervised learning; one-vs-rest nonlinear support vector machine models with recursive feature elimination |
Limitations |
|
|
-Validation cohort was not available. |
|
|
- Needs to be further validated in a different population. |
|
Kruse et al. (2017) [16] (Bone and mineral disorders) |
- Retrospective data from 10,775 subjects from the national Danish patient database with information on DXA scans, medication reimbursements, healthcare use, and comorbidities of female subjects |
- Aim: to detect patient clusters with a high risk of fracture using an unsupervised clustering algorithm based on DXA scans, medication, and health care claims dataset |
Key implications |
|
- Reference label: not applicable |
- Unsupervised clustering identified four high risk clusters and two low-risk clusters among nine clusters, which had different patterns of medication usage, compliance, and clinical outcomes despite similar DXA results. |
|
- Comparator: not applicable |
- May provide novel insights into establishing indications for DXA screening. |
|
- Methods: unsupervised learning; Ward's-based hierarchical agglomerative clustering |
Limitations |
|
- Potential temporal changes in pharmacological treatment pattern during the 15-year observation period |
|
- Inherent limitations of the secondary use of a claims dataset; could not ascertain actual consumption of medication by individual subjects. |
|
Risk prediction |
Segar et al. (2019) [17] (Diabetes and related disorders) |
- 8,756 Patients without heart failure at baseline from the ACCORD trial dataset (50% training set; 50% internal validation set; conducted between 1999 to 2009) |
- Aim: to develop an ML model to predict incident heart failure among patients with type 2 diabetes |
Key implications |
|
- External validation set: 10,819 participants without prevalent heart failure from the ALLHAT trial |
- Reference label: incident hospitalization or death due to heart failure (captured and adjudicated by two independent reviewer physicians during the trial) |
- The ML-based models showed modest performance in prediction for incident heart failure among patients with type 2 diabetes in the external validation set (C-index 0.70 to 0.74). |
|
- Comparator: not applicable |
- Each 1-unit increment in the WATCH-DM score was associated with a 24% higher relative risk of heart failure within 5 years. |
|
- Methods: supervised learning; random survival forest-based model |
- Strength of analyzing a large number of participants from a well-phenotyped clinical trial population |
|
|
Limitations |
|
|
- Discrimination for heart failure with preserved ejection fraction was relatively low in the subgroup analysis. |
|
|
- Temporal changes of heart failure biomarkers and medications could not be reflected in the model. |
|
|
- Need to validate the model in lower-risk cohorts of individuals with type 2 diabetes. |
|
Su et al. (2019) [18] (Bone and mineral disorders) |
- 5,977 Community-dwelling American men aged 65 or older (MrOS cohort) with 10-year follow-up data |
- Aim: to develop a risk classification model for hip fracture prediction in community-dwelling men |
Key implications |
|
- Internal validation with 10-fold cross-validation |
- Reference label: incident hip fracture validated by a centralized physician using radiology reports or X-rays |
- Simple CART model with age and bone density showed similar performance in predicting incident hip fracture compared to the FRAX risk estimator as the current standard (AUROC 0.71 vs. AUROC 0.70). |
|
- Comparator: FRAX hip fracture risk >3.0% |
- Simple classification by age and BMD may have a similar predictive performance to the FRAX hip fracture risk category. |
|
- Methods: supervised learning; classification and regression tree (CART) analysis |
Limitations |
|
|
- Potential of overfitting |
|
|
- Limited statistical power for comparison of discrimination statistics due to low incidence of hip fracture. |
|
Basu et al. (2018) [19] (Diabetes and related disorders) |
- 10,251 ACCORD trial participants aged 40 to 79 years with type 2 diabetes, HbA1c 7.5% or higher, or cardiovascular diseases or risk factors, those who randomized to target HbA1c <6.0% (intensive) vs. 7.0%–7.9% (standard group) |
- Aim: to identify subgroups with a heterogeneous treatment effect in response to intensive glycemic therapy |
Key implications |
|
- Reference label: treatment effect defined as the absolute difference in the all-cause mortality rate between the intensive and standard therapy groups |
- Compared to 3.7% increased mortality by intensive vs. standard therapy in group 4, group 1 showed a 2.3% mortality reduction in the intensive therapy group (95% CI, –0.2% to 4.5%), which made the obvious contrast with the main result from the study. |
|
- Comparator: not applicable |
- Identified characteristics of patients who may have benefited from intensive glycemic therapy (younger individuals with relatively low hemoglycosylation index) |
|
- Methods: supervised learning; gradient forest analysis |
- Offered an example to find, clinically meaningful subgroups with heterogeneous treatment effects using data from randomized trials. |
|
Limitations |
|
- Post hoc analysis of a single trial that was conducted before the development of recent diabetes medications with cardiovascular benefits. |
|
Fan et al. (2019) [20] (Pituitary diseases) |
- Retrospective cohort of 668 patients with acromegaly included age, gender, hypertension, blood glucose, laboratory values, maximal tumor diameter, bilateral Knosp grade based on magnetic resonance imaging findings, and surgical methods |
- Aim: to develop an ML model for preoperative prediction of transsphenoidal surgery response in patients with acromegaly |
Key implications |
|
- Internal validation set (n=134) |
- Reference label: remission (at 3 months after surgery, either nadir growth hormone <4 ng/mL after oral glucose tolerance test or GH <1.0 ng/mL in a random sample with normal IGF-1 levels) |
- The ML model predicted remission after surgery better than standard Knosp grade (AUROC 0.82 vs. 0.71). |
|
- Comparator: Knosp grade |
- Showed an exemplary case of applying various types of ML algorithms in endocrine diseases with relatively low frequency. |
|
- Methods: supervised learning; random forest, logistic regression, logistic generalized additive models, gradient boosting decision tree, gradient boosting decision tree, adaptive boosting, extreme gradient boost model |
Limitations |
|
|
- Single-center study |
|
|
- Limited by short study follow-up duration (remission determined at 3 months) |
|
|
- Omitted radiomics features |
|
Zaborek et al. (2019) [21] (Thyroid diseases) |
- Retrospective cohort of 598 patients who underwent total or completion thyroidectomy with pathology showing benign thyroid disease |
- Aim: to develop an ML-based levothyroxine dosing scheme after total thyroidectomy to achieve euthyroidism |
Key implications |
|
- Initiated levothyroxine at 1.6 μg/kg/day, with subsequent dose titration at 6- to 8-week intervals |
- Reference label: electronic health record-based euthyroid dosing |
- The predictive accuracy of the dose-suggestion algorithm was modest (64.8%), which was better than standard weight-based dosing (51.3%). |
|
- Internal validation with 10-fold cross-validation |
- Comparator: standard weight-based dosing |
- Provided an ML algorithm to suggest dosing scheme of levothyroxine after total thyroidectomy, with better accuracy across body mass index levels |
|
- Methods: supervised learning; support vector machine, Bayesian recurrent neural network, decision trees, random forests, ordinary least squares regression, Poisson regression, gamma regression, ridge regression, LASSO |
Limitations |
|
|
- Limited to dataset from a single institution; need further validation in an external dataset |
|
|
- Missing information regarding genetic factors and drug compliance; may hinder applicability to the real-world setting. |
|
Oroojeni Mohammad Javad et al., (2019) [5] (Diabetes and related disorders) |
- Medical records of 87 patients with type 1 diabetes from Mass General Hospital; data for each patient's visits over a 10-year period (training set) between 2003 to 2013; HbA1c, body mass index, activity level, alcohol usage status, insulin (Lantus) dose |
- Aim: to explore an effective reinforcement learning framework for determining the optimal long-acting insulin dose for patients with type 1 diabetes |
Key implications |
|
- External validation with 60 cases |
- Reference label: physician-prescribed insulin dose |
- The physician-prescribed insulin dose was within the dosing interval recommended by the Q-learning algorithm in 88% of test cases. |
|
- Comparator: not applicable |
- A proof-of-concept study to provide clinical decision support for determining insulin dose in patients with type 1 diabetes, by applying reinforcement learning algorithm |
|
- Methods: reinforcement learning; Q-learning with reward function set from HbA1c status at the visit and change of HbA1c from the past visit |
Limitations |
|
|
- Limited by omitting lifestyle information regarding diet, stress, and medication adherence |
|
|
- A relatively small training set |
|
|
- Only one type of insulin (Lantus) was examined in the model. |
|
Translational research Liu et al. (2020) [22] (Diabetes and related disorders) |
- 20 Drug-naive individuals with prediabetes (discovery cohort) |
- Aim: to find an ML model for predicting exercise responsiveness determined from exercise-induced alterations in the gut microbiota |
Key implications |
|
- Determined exercise responders and non- responders after 12-week high-intensity exercise training |
- Reference label: responders defined as a decrease in the homeostatic model assessment of insulin resistance greater than two-fold technical error |
- The ML model identified 14 microbiome species and 15 metabolites from human feces were able to predict exercise responsiveness (AUROC 0.75 in the validation set). |
|
- Collected pre- and post- exercise period feces to analyze gut microbiota profile |
- Comparator: not applicable |
- Provide an example of applying ML principles to human-to-mice translational study based on microbiome dataset |
|
- Internal validation with 10-fold cross-validation |
- Methods: supervised learning; random forest model |
Limitations |
|
|
- Relatively small sample size |
|
|
- Limited to Chinese males only |
|
|
- Need further validation in different population set |
|
Williams et al. (2019) [23] (Miscellaneous) |
- Prospectively collected data from archived samples, clinical data, with approximately 85 million protein measurements in 16,894 participants from various cohorts including UK Whitehall II, Fenland, HUNT3, US Covance, HERITAGE Family studies |
- Aim: to develop plasma protein-phenotype models for 11 different health indicators (focusing on percentage body fat and incident cardiovascular events as outcomes) |
Key implications |
|
- 70% Derivation set (with five repeats of 10-fold cross-validation), 15% refinement set, and 15% validation set for large (thousands) cohort |
- Reference label: percentage body fat measured by DXA; incident cardiovascular events ascertained in each cohort |
- The ML algorithm found proteins associated with body fat percentage (leptin, FABP, SFRP4) and CV events (gelsolin, antithrombin III, sTREM-1). |
|
- 80% Derivation set (with 10-fold cross-validation); 20% validation set for smaller dataset (hundreds) |
- Comparator: not applicable |
- Reveals the potential of ML algorithm application to find novel proteomics-based biomarkers in large-scale, well-established cohorts. |
|
- Methods: supervised learning; dimensionality reduction by false-recovery rate-corrected P values, proportional hazards elastic net models |
Limitations |
|
|
- Caucasian bias in some cohorts; may not be generalizable to different populations. |
|
|
- Need future investigation for examining the sensitivity of current research findings for longitudinal changes in health status or risks |
|
Shomorony et al. (2020) [6] (Miscellaneous) |
- 1,385 Data features using a multimodal dataset collected from 1,253 individuals including data of whole- genome sequencing, microbiome sequencing, global metabolome, insulin resistance, whole body and brain magnetic resonance imaging, bone densitometry, computed tomography scans, routine clinical laboratory tests, family history of disease and medication, and anthropometric measurements |
- Aim: to identify multimodal biomarker signatures of health and disease risk using the unsupervised approach |
Key implications |
|
- External validation set: 1,083 individuals from a separate cohort (TwinsUK registry) |
- Reference label: not applicable |
- 1-stearoyl-2-dihomo-linolenoyl-GPC and 1-(1-enyl-palmitoyl)-2-oleoyl-GPC were identified as novel biomarkers for diabetes, whereas cinnamoylglycine showed a novel association with lean mass percentage. |
|
- Comparator: not applicable |
- Provided an example of applying unsupervised learning algorithms to find novel associations and biomarker signatures associated with health and disease statues in a large, multimodal dataset |
|
- Methods: unsupervised learning; Louvain community detection, graphical LASSO for network analysis, Markov network analysis |
Limitations |
|
|
- Underpowered to detect the effects of polygenic risk scores based on common variants for certain traits (explaining a relatively small fraction of the phenotypic variance) |




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download