Abstract
Anaplastic thyroid cancer (ATC) is a lethal human cancer with a 5-year survival rate of less than 10%. Recently, its genomic and transcriptomic characteristics have been extensively elucidated over 5 years owing to advance in high throughput sequencing. These efforts have extended molecular understandings into the progression mechanisms and therapeutic vulnerabilities of aggressive thyroid cancers. In this review, we provide an overview of genomic and transcriptomic alterations in ATC and poorly-differentiated thyroid cancer, which are distinguished from differentiated thyroid cancers. Clinically relevant genomic alterations and deregulated signaling pathways will be able to shed light on more effective prevention and stratified therapeutic interventions for affected patients.
Thyroid cancer is one of the most common malignancy in human endocrine systems [1]. The most prevalent types of thyroid cancer are differentiated thyroid cancers (DTCs) which are developed from follicular cells of thyroid [2]. Papillary thyroid cancer (PTC; 80% to 85%) and follicular thyroid cancer (FTC; 10% to 15%) account for the majority of thyroid cancers and most patients have good prognosis [23]. However, patients with advanced forms of thyroid cancer including poorly-differentiated thyroid cancer (PDTC) and anaplastic thyroid cancer (ATC) show worse prognosis compared with DTC [4]. Among them, ATC is the most fatal human cancer type with a median survival of 5 months but there is no effective therapeutic option for this fatal disease yet [4].
Through the recent surge of next-generation sequencing (NGS) technology, a myriad number of studies have been successfully deciphered the underlying molecular nature of human cancers [56]. Since various genetic factors affect therapeutic vulnerability or resistance to drugs [789], understanding molecular events occurred during thyroid cancer progression would provide effective therapeutic strategy. Also, the early detection of those markers in DTC is important to prevent and predict the progression. In this review, we focus on key genomic and transcriptomic events in aggressive thyroid cancers discovered in NGS era.
BRAF is a proto-oncogene encoding a serine/threonine-protein kinase and altered in 69% to 71% of PTC (classical type) [1011]. This gene has a crucial role in the Ras/Raf/mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) pathway and its alterations affect cell cycle and progression [12]. The Ras oncogene family (hereafter referred as RAS) including HRAS, KRAS, and NRAS is most commonly altered in follicular-patterned thyroid tumors (follicular adenoma [FA], FTC, and follicular variant of PTC) [101113]. Since BRAF and RAS are tightly associated with the histology of thyroid cancer, they could be identifiers for tumor origin of ATC.
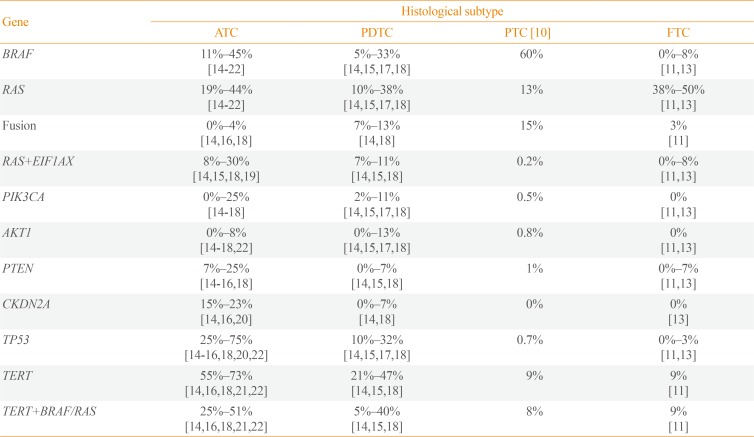
In ATC, BRAF and RAS mutations were identified with frequencies of 11%–45% and 19%%44%, respectively (Table 1) [141516171819202122]. Those alterations usually displayed similar incidence or sometimes BRAF was slightly more frequent than RAS [141516171819]. There is one study describing exceptionally high frequency of BRAF (91%) [23], and a few studies reported that RAS was more common than BRAF [172122]. In general, the combined frequency of BRAF and RAS mutations accounts for more than 50% of ATCs which signifies that ATC is commonly originated from DTC rather than de novo [14151618192223]. It is also reported that 5%–33% and 10%–38% of PDTCs harbor BRAF and RAS mutations, respectively [1418].
It has been reported that ATC is barely developed with fusion gene. According to Landa et al. [18], only one out of 33 ATCs had a fusion gene, but 13% of PDTC had well-known thyroid cancer driver fusions (five RET, three paired box 8 [PAX8]-peroxisome proliferator activated receptor gamma [PPARG], and three ALK receptor tyrosine kinase [ALK] fusions). In PDTC, ALK rearrangement is known to be relatively common; 16% of thyroid tumors with ALK rearrangement are PDTC [2425]. Intriguingly, striatin (STRN)-ALK is predominantly found in thyroid cancer including PDTC rather than EMAP like 4 (EML4)-ALK, the most well-known ALK rearrangement. A recent in vivo study showed that PDTC is frequently developed in 22% and 36% of thyroglobulin (Tg)-STRN-ALK mice with and without goitrogen treatment, respectively [26]. In a study with the largest cohort of 196 ATCs, only 4% of ATCs harbored fusion genes including three BRAF (two with KIAA1549 and one with epidermal growth factor receptor pathway substrate 15 like 1 [EPS15L1]), two neurotrophic receptor tyrosine kinase 1 (NTRK1) (with lamin A/C [LMNA] and tropomyosin 3 [TPM3]), and three coiled-coil domain containing 6 (CCDC6)-RET fusions [16]. The low prevalence of fusion gene in ATC is also confirmed by genomic profiling of thyroid cancer cell lines. There were only two out of 31 ATC cell lines with oncogenic fusions including makorin ring finger protein 1 (MKRN1)-BRAF (in THJ1-6T) and fibroblast growth factor receptor 2 (FGFR2)-oxoglutarate dehydrogenase (OGDH) (in THJ-29T) [27].
Eukaryotic translation initiation factor 1A X-linked (EIF1AX) encodes an essential eukaryotic translation initiation factor and is recently proposed as a cancer driver gene. Its role in driving cancer is firstly reported in uveal melanoma [28]. EIF1AX is usually co-mutated with G protein subunit alpha q (GNAQ) or G protein subunit alpha 11 (GNA11), and melanoma patients with EIF1AX mutations have good prognosis compared with others [29]. From pan-cancer data, only 0.3% of tumors (33/10,967) harbor hotspot mutations in EIF1AX (from http://www.cbioportal.org) [30]. Until now, its role in human cancer is not fully investigated, but in vitro analysis showed that increased EIF1AX activity triggers protein translation and cell proliferation [31]. In The Cancer Genome Atlas (TCGA) study, EIF1AX was confirmed as a driver gene of PTC (mostly for follicular variant types) [10]. It is also altered in FA and minimally invasive FTC [1113], which signifies less aggressive nature of EIF1AX. In contrast to uveal melanoma, EIF1AX mutation is sole event and does not cooperate other mutation in DTC. It is mutually exclusive with other driver mutations such as BRAF, RAS, and fusion genes [101113].
However, the mutually exclusiveness of EIF1AX to other driver genes is weaken and it is often co-occurred with RAS in aggressive thyroid cancers. According to Kunstman et al. [19], all of tumors with EIF1AX also harbored RAS mutations (NRAS or KRAS), and it accounts for 50% of RAS-positive ATCs. Furthermore, several studies with the larger cohorts confirmed the prevalence of EIF1AX+RAS in PTC, widely invasive FTC (wiFTC), PDTC, and ATC as 0.2%, 17%, 7%–11%, and 8%–30%, respectively [10141518]. In particular, EIF1AX-A113splice variant was commonly identified in EIF1AX-RAS co-mutated tumors [141832]. A recent experimental analysis suggested that protein synthesis in EIF1AX-A113splice knock-in thyroid cancer cell lines is increased through an activating transcription factor 4 (ATF4)-induced dephosphorylation of eukaryotic initiation factor 2 alpha (EIF2α) [32]. Furthermore, EIF1AX promotes the mammalian target of rapamycin (mTOR) activation to amino acid supply through cooperation of ATF4 and cellular myelocytomatosis oncogene (c-MYC). This study also showed that combinational treatment of mTOR kinase inhibitor (AZD8055) with either MEK inhibitor (trametinib) or bromodomain-containing protein 4 (BRD4) inhibitor (JQ1) to EIF1AX-A113splice knock-in CAL62 cell line resulted in huge tumor reduction and decreased c-MYC and mTOR protein levels.
Meanwhile, cyclin E1 (CCNE1) amplification rather than EIF1AX mutation was also reported to be occurred with 4% of RAS-positive ATCs [1620]. In those reports, targeted sequencing platform from Foundation Medicine Inc. was applied which does not include EIF1AX for genomic profiling. Therefore, the implication of CCNE1 and its relationship to EIF1AX or RAS is needed to be further investigated.
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) and AKT serine/threonine kinase 1 (AKT1) are members of the oncogenic phosphoinositide 3-kinase (PI3K)/AKT/mTOR signaling pathway which are altered in diverse human malignancies [33]. The prevalence of PIK3CA mutations in human cancer is varied across cancer types: e.g., endometrial carcinoma (37%), breast cancer (31%), colorectal carcinoma (17%), pancreatic carcinoma (3%), and melanoma (2%) [34]. On the other hand, AKT1 is less frequently altered in human cancers relative to PIK3CA, and most commonly mutated in adenoid cystic carcinoma (4%), endometrial carcinoma (3%), and breast cancer (3%) [34]. In PTC, the prevalence of PIK3CA and AKT1 mutations are known to be 0.5% and 0.8%, respectively [10], and they are not reported in follicular thyroid tumors [1113]. Moreover, they are slightly frequent in metastatic PTC (3% for both) compared with PTC [14]. However, several reports showed that PIK3CA and AKT1 are frequently altered in aggressive thyroid cancers. The frequency of PIK3CA mutation in PDTC and ATC is 2%–11% and 0%–25%, respectively [1415161718], and AKT1 (especially for E17K) is found in 0%–13% and 0%–8% of PDTC and ATC, respectively [1415161718]. It has been reported that PIK3CA mutations are frequently co-occurred with BRAF or RAS in diverse types of advanced cancers [353637]. However, PIK3CA mutations are usually reported as co-existed with BRAF in aggressive thyroid cancers [1415161718192338], and only a few studies showed that RAS-PIK3CA co-mutation is more frequent than BRAF-PIK3CA co-mutation [2122]. In agreement with other cancer types, AKT1E17K is less common compared with PIK3CA, but it is also co-occurred with BRAF [141622]. Moreover, PIK3CA and AKT1 mutations are completely mutually exclusive to each other as reported in other cancer types [39]. It suggests that those mutations may have biologically equivalent role in activating PI3K/AKT/mTOR pathway to drive progression of thyroid cancer [4041].
Furthermore, high prevalence of phosphatase and tensin homolog (PTEN) alteration (7% to 25%) which regulates PI3K/AKT/mTOR signaling pathway was also reported in aggressive thyroid cancers [1415161820222342]. Notably, two studies reported that most of PTEN alterations (45%, 10/22) was co-occurred with neurofibromin 1 (NF1) or RB transcriptional corepressor 1 (RB1) rather than BRAF or RAS [1618].
Telomerase activation is one of the hallmarks of cancer [43]. The activation of telomerase reverse transcriptase (TERT) is usually induced by the mutations in the promoter region known as C228T or C250T (96% in pan-cancer) [44]. In DTC, several reports confirmed its significance in poor survival of the patients [454647]. Especially, when TERT promoter mutations co-exist with BRAFV600E or RAS mutations, they exert a negative synergistic effect on prognosis. Several studies demonstrated the potential mechanism of this synergism between the mutations; BRAFV600E-induced E26 transformation-specific or E-twenty-six (ETS) transcription factors increase TERT expression by binding to the ETS-binding site generated by TERT promoter mutation [4849]. The frequency of TERT promoter mutations in PTC are reported as 9% [10], but their incidences are greatly increased in metastatic PTC (60%) [44]. Moreover, 21% to 47% PDTC and 55% to 73% ATC are known to have TERT promoter mutation that again emphasize the critical role of TERT in thyroid cancer progression [141516182122]. Recently Yoo et al. [14] reported the novel types of TERT alterations in wiFTC including fusion gene and translocation in TERT upstream, and they also induced extremely high expression levels of TERT. The rearrangements with/within TERT are often identified in aggressive human cancer types [505152]. Therefore, TERT rearrangements in thyroid cancer are also significant events that can induce the progression. Moreover, previously analyzed aggressive thyroid cancers without TERT promoter mutations more likely to harbor unidentified TERT rearrangements since these events are mutually exclusive to TERT promoter mutations and most studies did not focus on intergenic region [1453].
Tumor protein p53 (TP53) is the most well-studied human tumor suppressor and its incidence is reported as 37% in pan-cancer data (3,786/10,225) [54]. More than 90% of patients in some cancer types, such as ovarian cancer and uterine carcinosarcoma, harbor mutation in TP53, but the seven cancer types have incidence less than 5% (0.7% for DTC) [1011]. Mutations in this gene is tightly associated with the progression and poor prognosis of human cancers including ATC [55565758]. In contrast to DTC, its prevalence is extremely increased according to the aggressiveness of thyroid cancer; the frequency of TP53 in PDTC and ATC were reported as 10%–32% and 25%–75%, respectively [14151617182122]. Since TP53 alterations are relatively rare in metastatic PTC (13%) and wiFTC (8%) which are early advanced forms of DTC whereas TERT promoters are commonly mutated in them (48% and 75% for each), it is suspected that TERT and TP53 are major players triggering early and late progression of thyroid cancer, respectively (Fig. 1) [14].
Cyclin dependent kinase inhibitor 2A (CDKN2A) encodes a tumor suppressor protein, p16INK4a, which has a critical role in the inhibition of cell cycle progression [59]. This gene is located at 9q21.3 locus and is known to be deactivated by diverse mechanisms such as homozygous loss, exon skipping, truncating mutations, and epigenetic silencing [60]. In particular, loss of CDKN2A and its significance in cancer progression have been well reported in several types of cancers [616263]. However, its association to thyroid cancer progression is recently proposed by a few studies. There are three reports that discovered CDKN2A loss in aggressive thyroid cancers, and its incidence in ATC was reported as 15% to 23% [141620]. In addition, CDKN2A deletion is also frequent in thyroid cancer cell lines (47%) [27]. It could be robustly associated with thyroid cancer progression since this type of variation is not identified in DTC [1013]. According to Yoo et al. [14], CDKN2A loss is not only associated with the progression of ATC, but also involved in poor prognosis and survival of patient with advanced DTC and ATC. By thyroid differentiated score (TDS) analysis from TCGA [10], the poorer differentiation status in ATCs with CDKN2A loss relative to those with CDKN2A2n was identified. Furthermore, patients with ATC or advanced DTCs harboring CDKN2A/p16 loss showed poor disease-specific survival. Hence, CDKN2A can be used as a useful biomarker to monitor the prognosis of thyroid cancer patients.
In addition to mutation profiles, some reports described the additional genomic features related to aggressive thyroid cancers. A recent study reported that one out of eleven ATCs showed tendency to microsatellite instability high (MSI-H) [64]. There is a report that also showed MSI in the synchronous FTC (MSI borderline), PDTC, and ATC from one patient [65]. Moreover, some PDTC (T243 with MutS homolog 2 [MSH2] mutation) and FTC (FTC-133, FTC-236, and FTC-238 with MSH6, MSH6/MLH1, and MSH6 mutations, respectively) cell lines have mutations in mismatch repair genes and exceptionally high tumor mutational burden (TMB) [27]. Since patients with MSI-H, mismatch repair deficiency (MMR-D), or high TMB tumors show the durable clinical benefit from immunotherapy [66676869], this therapy could be a new option for the minor portion of patients with aggressive thyroid cancer. Also, the mutational signature analysis from two studies showed increased Apolipoprotein B mRNA Editing Catalytic Polypeptide-like (APOBEC) cytidine deaminase activity in aggressive thyroid cancers [1416]. As an APOBEC-related mutagenesis also leads to high TMB and linked to the immune activations [7071], patients with this mutational signature would be also potential target of immunotherapy.
Until now, only a small number of studies investigated transcriptomic nature of aggressive thyroid cancers. Landa et al. [18] showed the transcriptomic feature of ATC using TCGA's BRAFV600E-RAS score (BRS) analysis [10]. From the original investigation of TCGA, DTC could be sub-classified into BRAFV600E-like (with negative BRS) and RAS-like (with positive BRS) (Fig. 2A). In general, BRAFV600E-like and RAS-like are associated with BRAF and RAS mutations, respectively, and BRAFV600E-like DTCs display more aggressive clinical characteristics than RAS-like DTCs [11]. However, ATC did not follow the BRS rules, but most of them showed BRAFV600E-like signature (negative BRS) although some of them harbored RAS mutations [18]. Yoo et al. [14] also reported a consistent result using BRS analysis (negative BRS for both BRAF and RAS mutated ATCs) and also proposed advanced view of transcriptomic feature. Based on principal component analysis, BRAF-positive and RAS-positive ATCs showed similar expression profile as in BRS analysis, but they were grouped into novel molecular subtype, ATC-like, rather than BRAFV600E-like. This result signifies that ATCs share similar global transcriptomic features regardless of their mutational status and have totally distinctive molecular characteristics
from DTC.
There are two studies conducted on differentially expressed genes (DEGs) and pathways analyses in ATC (Fig. 2B). Kasaian et al. [72] identified that extracellular matrix (ECM)-receptor interaction, focal adhesion, cell cycle, p53, and general cancer signaling pathways were up-regulated in ATC correspondence to PTC and normal thyroid tissue [72]. Moreover, cancer-related genes such as MYC, MTOR, protein kinase C alpha (PRKCA), and transforming growth factor beta 1 (TGFB1) were overexpressed. Meanwhile, tight junctions, cell adhesion molecules, various metabolism pathways, and thyroid differentiation signature genes such as TG, transcription termination factor 1 (TTF1), thyroid stimulating hormone receptor (TSHR), and thyroid peroxidase (TPO) were decreased. Yoo et al. [14] also discovered similar transcriptome changes in ATC. DEG and pathway analyses were performed separately using ATC and DTC based on driver mutation status (BRAF and RAS). This study showed that both BRAF-positive and RAS-positive ATCs displayed up-regulation of the mitogen-activated protein kinase (MAPK), ECM-receptor, focal adhesion, cell cycle, p53, and general cancer signaling pathways as Kasaian et al. [72] reported. Also, BRAF-positive and RAS-positive ATCs showed activation of vascular endothelial growth factor (VEGF)/notch and Janus kinase (JAK)-Signal transducers and activators of transcription (STAT) signaling pathways, respectively. In particular, suppressor of cytokine signaling 3 (SOCS3), BCL2 like 1 (BCL2L1), and MYC were up-regulated in RAS-positive ATCs, and in vitro experiments showed that their expression level and cell proliferation were decreased upon treatment of ruxolitinib, JAK inhibitor. Furthermore, several metabolism pathways and most of thyroid differentiation genes included in TDS analysis were down-regulated in ATC.
Understanding immune cell signature in human cancer is one of the important field in cancer biology since they are closely involved in the cancer progression or therapeutic response [73]. Also, the high degree of immune cell infiltration is reported to be associated with unfavorable clinical outcomes of patients with DTC [74]. Landa et al. [18] showed that ATC have elevated M2 macrophage infiltration compared with PDTC based on the expression profile of 68 genes which are subset of 78 genes associated with M2 macrophages [1875]. Giannini et al. [76] also displayed increased macrophage score in ATC compared with PDTC, PTC, and normal thyroid. Moreover, tumor infiltrating leukocyte, T-cells (CD8+ and exhausted CD8+ T-cells), and cytotoxicity cell scores were significantly elevated in ATC. Interestingly, aforementioned immune cell infiltrations were less active in PDTC relative to PTC. In addition, two types of immune signatures, ATC-like and PDTC-like, in thyroid cancer were found. They investigated four categories of immune signatures: hot, altered-immunosuppressed, altered-excluded, and cold [77]. According to the study, most of ATCs were defined as two main immune contextures, hot (34%; T-cell-inflamed) and altered-immunosuppressed (50%; a low degree of T-cell infiltration, but presence of soluble inhibitory mediators, immune suppressive cells, and T-cell checkpoints). On the other hand, PDTC showed another two immune contextures: cold (65%; non-T-cell-inflamed) and altered-excluded (14%; T-cell at the invasive margins or tumor edge without intratumoral infiltration).
In addition to immune cell infiltration, the up-regulation or amplification of CD274 (encodes programmed cell death 1 ligand 1 [PD-L1]) or PDCD1LG2 (encodes programmed cell death 1 ligand 2 [PD-L2]) which belong to the family of immune checkpoint proteins were reported in a handful of ATCs [1416]. There have been also various reports showed expression of PD-1 or PD-L1 in ATC using immunohistochemistry [78798081]. These markers are considered as imperfect predictor of immunotherapy response alone [82], but combination of their expression and other genomic features (MSI-H, MMR-D, or TMB) could be better predictor for immunotherapy response of patients with ATC.
Herein, we reviewed the recent findings in NGS era regarding the genomic and transcriptomic changes associated with ATC and PDTC. The key molecular features of ATC could be shortened as follows. First, co-mutations of TP53 (25% to 75%) as well as TERT (55% to 73%) are hallmarks of ATC considering their remarkably high prevalence. Second, the additional mutational hits in oncogenes such as PIK3CA/AKT1 (with BRAF) or EIF1AX (with RAS). Third, BRAF-positive and RAS-positive ATCs display similar global transcriptome features, ATC-like. Fourth, the extra activations of cancer related signaling pathways such as ECM-receptor interaction, focal adhesion, cell cycle, p53, and MAPK. At last, dysfunction of thyroid hormone and various metabolism signaling pathways.
Despite the extensive molecular profiling of thyroid cancers, only limited number of studies regarding PDTC was reported yet. In particular, none of literature performed comprehensive DEG and pathway analyses including PTDC as well as ATC and DTC. Based on the findings from Giannini et al. [76], there might be dynamic molecular changes rather than linear progression as histological grades from DTC through PDTC to ATC. Hence, investigating this molecular feature would allow us to deeper understanding of molecular pathogenesis of thyroid cancer and improve patient care.
ACKNOWLEDGMENTS
This study was funded by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Science, ICT & Future Planning (NRF-2019R1A2C2084332).
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; 69:7–34. PMID: 30620402.

2. Dralle H, Machens A, Basa J, Fatourechi V, Franceschi S, Hay ID, et al. Follicular cell-derived thyroid cancer. Nat Rev Dis Primers. 2015; 1:15077. PMID: 27188261.

3. La Vecchia C, Malvezzi M, Bosetti C, Garavello W, Bertuccio P, Levi F, et al. Thyroid cancer mortality and incidence: a global overview. Int J Cancer. 2015; 136:2187–2195. PMID: 25284703.

4. Ibrahimpasic T, Ghossein R, Shah JP, Ganly I. Poorly differentiated carcinoma of the thyroid gland: current status and future prospects. Thyroid. 2019; 29:311–321. PMID: 30747050.

5. Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017; 168:584–599. PMID: 28187282.

6. Song YS, Park YJ. Genomic characterization of differentiated thyroid carcinoma. Endocrinol Metab (Seoul). 2019; 34:1–10. PMID: 30912334.

7. Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012; 11:873–886. PMID: 23060265.

8. Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010; 363:1693–1703. PMID: 20979469.
9. Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Bergqvist S, Brooun A, et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016; 374:54–61. PMID: 26698910.
10. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014; 159:676–690. PMID: 25417114.
11. Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho H, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PLoS Genet. 2016; 12:e1006239. PMID: 27494611.

12. Mercer KE, Pritchard CA. Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim Biophys Acta. 2003; 1653:25–40. PMID: 12781369.

13. Jung SH, Kim MS, Jung CK, Park HC, Kim SY, Liu J, et al. Mutational burdens and evolutionary ages of thyroid follicular adenoma are comparable to those of follicular carcinoma. Oncotarget. 2016; 7:69638–69648. PMID: 27626165.

14. Yoo SK, Song YS, Lee EK, Hwang J, Kim HH, Jung G, et al. Integrative analysis of genomic and transcriptomic characteristics associated with progression of aggressive thyroid cancer. Nat Commun. 2019; 10:2764. PMID: 31235699.

15. Duan H, Li Y, Hu P, Gao J, Ying J, Xu W, et al. Mutational profiling of poorly differentiated and anaplastic thyroid carcinoma by the use of targeted next-generation sequencing. Histopathology. 2019; 75:890–899. PMID: 31230400.

16. Pozdeyev N, Gay LM, Sokol ES, Hartmaier R, Deaver KE, Davis S, et al. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res. 2018; 24:3059–3068. PMID: 29615459.

17. Chen H, Luthra R, Routbort MJ, Patel KP, Cabanillas ME, Broaddus RR, et al. Molecular profile of advanced thyroid carcinomas by next-generation sequencing: characterizing tumors beyond diagnosis for targeted therapy. Mol Cancer Ther. 2018; 17:1575–1584. PMID: 29695638.

18. Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016; 126:1052–1066. PMID: 26878173.

19. Kunstman JW, Juhlin CC, Goh G, Brown TC, Stenman A, Healy JM, et al. Characterization of the mutational landscape of anaplastic thyroid cancer via whole-exome sequencing. Hum Mol Genet. 2015; 24:2318–2329. PMID: 25576899.

20. Khan SA, Ci B, Xie Y, Gerber DE, Beg MS, Sherman SI, et al. Unique mutation patterns in anaplastic thyroid cancer identified by comprehensive genomic profiling. Head Neck. 2019; 41:1928–1934. PMID: 30758123.

21. Tiedje V, Ting S, Herold T, Synoracki S, Latteyer S, Moeller LC, et al. NGS based identification of mutational hotspots for targeted therapy in anaplastic thyroid carcinoma. Oncotarget. 2017; 8:42613–42620. PMID: 28489587.

22. Bonhomme B, Godbert Y, Perot G, Al Ghuzlan A, Bardet S, Belleannee G, et al. Molecular pathology of anaplastic thyroid carcinomas: a retrospective study of 144 cases. Thyroid. 2017; 27:682–692. PMID: 28351340.

23. Jeon MJ, Chun SM, Kim D, Kwon H, Jang EK, Kim TY, et al. Genomic alterations of anaplastic thyroid carcinoma detected by targeted massive parallel sequencing in a BRAF (V600E) mutation-prevalent area. Thyroid. 2016; 26:683–690. PMID: 26980298.
24. Kelly LM, Barila G, Liu P, Evdokimova VN, Trivedi S, Panebianco F, et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci U S A. 2014; 111:4233–4238. PMID: 24613930.

25. Panebianco F, Nikitski AV, Nikiforova MN, Kaya C, Yip L, Condello V, et al. Characterization of thyroid cancer driven by known and novel ALK fusions. Endocr Relat Cancer. 2019; 26:803–814. PMID: 31539879.

26. Nikitski AV, Rominski SL, Wankhede M, Kelly LM, Panebianco F, Barila G, et al. Mouse model of poorly differentiated thyroid carcinoma driven by STRN-ALK fusion. Am J Pathol. 2018; 188:2653–2661. PMID: 30125543.

27. Landa I, Pozdeyev N, Korch C, Marlow LA, Smallridge RC, Copland JA, et al. Comprehensive genetic characterization of human thyroid cancer cell lines: a validated panel for preclinical studies. Clin Cancer Res. 2019; 25:3141–3151. PMID: 30737244.

28. Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013; 45:933–936. PMID: 23793026.

29. Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017; 32:204–220. PMID: 28810145.

30. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. PMID: 22588877.
31. Yu C, Luo C, Qu B, Khudhair N, Gu X, Zang Y, et al. Molecular network including eIF1AX, RPS7, and 14-3-3γ regulates protein translation and cell proliferation in bovine mammary epithelial cells. Arch Biochem Biophys. 2014; 564:142–155. PMID: 25281768.

32. Krishnamoorthy GP, Davidson NR, Leach SD, Zhao Z, Lowe SW, Lee G, et al. EIF1AX and RAS mutations cooperate to drive thyroid tumorigenesis through ATF4 and c-MYC. Cancer Discov. 2019; 9:264–281. PMID: 30305285.

33. Dienstmann R, Rodon J, Serra V, Tabernero J. Picking the point of inhibition: a comparative review of PI3K/AKT/mTOR pathway inhibitors. Mol Cancer Ther. 2014; 13:1021–1031. PMID: 24748656.

34. Millis SZ, Ikeda S, Reddy S, Gatalica Z, Kurzrock R. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors. JAMA Oncol. 2016; 2:1565–1573. PMID: 27388585.
35. Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, et al. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther. 2012; 11:485–491. PMID: 22135231.

36. Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011; 6:e22769. PMID: 21829508.

37. Chiu JW, Krzyzanowska MK, Serra S, Knox JJ, Dhani NC, Mackay H, et al. Molecular profiling of patients with advanced colorectal cancer: princess margaret cancer centre experience. Clin Colorectal Cancer. 2018; 17:73–79. PMID: 29128266.

38. Gibson WJ, Ruan DT, Paulson VA, Barletta JA, Hanna GJ, Kraft S, et al. Genomic heterogeneity and exceptional response to dual pathway inhibition in anaplastic thyroid cancer. Clin Cancer Res. 2017; 23:2367–2373. PMID: 27797976.

39. Rudolph M, Anzeneder T, Schulz A, Beckmann G, Byrne AT, Jeffers M, et al. AKT1 (E17K) mutation profiling in breast cancer: prevalence, concurrent oncogenic alterations, and blood-based detection. BMC Cancer. 2016; 16:622. PMID: 27515171.

40. Bleeker FE, Felicioni L, Buttitta F, Lamba S, Cardone L, Rodolfo M, et al. AKT1(E17K) in human solid tumours. Oncogene. 2008; 27:5648–5650. PMID: 18504432.

41. Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007; 448:439–444. PMID: 17611497.
42. Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014; 4:64. PMID: 24782981.

44. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017; 23:703–713. PMID: 28481359.
45. Xing M, Liu R, Liu X, Murugan AK, Zhu G, Zeiger MA, et al. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol. 2014; 32:2718–2726. PMID: 25024077.

46. Melo M, da Rocha AG, Vinagre J, Batista R, Peixoto J, Tavares C, et al. TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J Clin Endocrinol Metab. 2014; 99:E754–E765. PMID: 24476079.

47. Song YS, Lim JA, Choi H, Won JK, Moon JH, Cho SW, et al. Prognostic effects of TERT promoter mutations are enhanced by coexistence with BRAF or RAS mutations and strengthen the risk prediction by the ATA or TNM staging system in differentiated thyroid cancer patients. Cancer. 2016; 122:1370–1379. PMID: 26969876.
48. Liu R, Zhang T, Zhu G, Xing M. Regulation of mutant TERT by BRAF V600E/MAP kinase pathway through FOS/GABP in human cancer. Nat Commun. 2018; 9:579. PMID: 29422527.

49. Song YS, Yoo SK, Kim HH, Jung G, Oh AR, Cha JY, et al. Interaction of BRAF-induced ETS factors with mutant TERT promoter in papillary thyroid cancer. Endocr Relat Cancer. 2019; 26:629–641. PMID: 30999281.

50. Diplas BH, He X, Brosnan-Cashman JA, Liu H, Chen LH, Wang Z, et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat Commun. 2018; 9:2087. PMID: 29802247.

51. Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015; 47:1411–1414. PMID: 26523776.

52. Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015; 526:700–704. PMID: 26466568.

53. Liang WS, Hendricks W, Kiefer J, Schmidt J, Sekar S, Carpten J, et al. Integrated genomic analyses reveal frequent TERT aberrations in acral melanoma. Genome Res. 2017; 27:524–532. PMID: 28373299.

54. Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated analysis of TP53 gene and pathway alterations in the Cancer Genome Atlas. Cell Rep. 2019; 28:1370–1384. PMID: 31365877.

55. Cho SY, Park C, Na D, Han JY, Lee J, Park OK, et al. High prevalence of TP53 mutations is associated with poor survival and an EMT signature in gliosarcoma patients. Exp Mol Med. 2017; 49:e317. PMID: 28408749.

56. Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014; 25:304–317. PMID: 24651012.

57. Acin S, Li Z, Mejia O, Roop DR, El-Naggar AK, Caulin C. Gain-of-function mutant p53 but not p53 deletion promotes head and neck cancer progression in response to oncogenic K-ras. J Pathol. 2011; 225:479–489. PMID: 21952947.

58. McFadden DG, Vernon A, Santiago PM, Martinez-McFaline R, Bhutkar A, Crowley DM, et al. p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer. Proc Natl Acad Sci U S A. 2014; 111:E1600–E1609. PMID: 24711431.

59. Liggett WH Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J Clin Oncol. 1998; 16:1197–1206. PMID: 9508208.

60. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012; 489:519–525. PMID: 22960745.
61. Bui NQ, Przybyl J, Trabucco SE, Frampton G, Hastie T, van de Rijn M, et al. A clinico-genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res. 2019; 9:12. PMID: 31528332.

62. Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 2015; 373:1926–1936. PMID: 26559571.

63. Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015; 160:1246–1260. PMID: 25748654.

64. Ravi N, Yang M, Gretarsson S, Jansson C, Mylona N, Sydow SR, et al. Identification of targetable lesions in anaplastic thyroid cancer by genome profiling. Cancers (Basel). 2019; 11:E402. PMID: 30909364.

65. Paulsson JO, Backman S, Wang N, Stenman A, Crona J, Thutkawkorapin J, et al. Whole-genome sequencing of synchronous thyroid carcinomas identifies aberrant DNA repair in thyroid cancer dedifferentiation. J Pathol. 2020; 250:183–194. PMID: 31621921.

66. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017; 357:409–413. PMID: 28596308.
67. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015; 348:124–128. PMID: 25765070.
68. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015; 372:2509–2520. PMID: 26028255.
69. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014; 371:2189–2199. PMID: 25409260.

70. Wang S, Jia M, He Z, Liu XS. APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. 2018; 37:3924–3936. PMID: 29695832.

71. Faden DL, Ding F, Lin Y, Zhai S, Kuo F, Chan TA, et al. APOBEC mutagenesis is tightly linked to the immune landscape and immunotherapy biomarkers in head and neck squamous cell carcinoma. Oral Oncol. 2019; 96:140–147. PMID: 31422205.

72. Kasaian K, Wiseman SM, Walker BA, Schein JE, Zhao Y, Hirst M, et al. The genomic and transcriptomic landscape of anaplastic thyroid cancer: implications for therapy. BMC Cancer. 2015; 15:984. PMID: 26680454.

73. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013; 14:1014–1022. PMID: 24048123.

74. Kim K, Jeon S, Kim TM, Jung CK. Immune gene signature delineates a subclass of papillary thyroid cancer with unfavorable clinical outcomes. Cancers (Basel). 2018; 10:E494. PMID: 30563160.

75. Coates PJ, Rundle JK, Lorimore SA, Wright EG. Indirect macrophage responses to ionizing radiation: implications for genotype-dependent bystander signaling. Cancer Res. 2008; 68:450–456. PMID: 18199539.

76. Giannini R, Moretti S, Ugolini C, Macerola E, Menicali E, Nucci N, et al. Immune profiling of thyroid carcinomas suggests the existence of two major phenotypes: an ATC-like and a PDTC-like. J Clin Endocrinol Metab. 2019; 104:3557–3575. PMID: 30882858.

77. Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019; 18:197–218. PMID: 30610226.

78. Cantara S, Bertelli E, Occhini R, Regoli M, Brilli L, Pacini F, et al. Blockade of the programmed death ligand 1 (PD-L1) as potential therapy for anaplastic thyroid cancer. Endocrine. 2019; 64:122–129. PMID: 30762153.

79. Chintakuntlawar AV, Rumilla KM, Smith CY, Jenkins SM, Foote RL, Kasperbauer JL, et al. Expression of PD-1 and PD-L1 in anaplastic thyroid cancer patients treated with multimodal therapy: results from a retrospective study. J Clin Endocrinol Metab. 2017; 102:1943–1950. PMID: 28324060.

80. Ahn S, Kim TH, Kim SW, Ki CS, Jang HW, Kim JS, et al. Comprehensive screening for PD-L1 expression in thyroid cancer. Endocr Relat Cancer. 2017; 24:97–106. PMID: 28093480.

81. Bastman JJ, Serracino HS, Zhu Y, Koenig MR, Mateescu V, Sams SB, et al. Tumor-infiltrating T cells and the PD-1 checkpoint pathway in advanced differentiated and anaplastic thyroid cancer. J Clin Endocrinol Metab. 2016; 101:2863–2873. PMID: 27045886.

82. Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019; 19:133–150. PMID: 30755690.

Fig. 1
The major genetic contributors to thyroid cancer progression. Progression mechanisms of BRAF-positive papillary thyroid cancer (PTC) and RAS-positive follicular thyroid cancer (FTC) are illustrated. TERT, telomerase reverse transcriptase; TP53, tumor protein p53; CDKN2A, cyclin dependent kinase inhibitor 2A; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; AKT1, AKT serine/threonine kinase 1; ATC, anaplastic thyroid cancer; EIF1AX, eukaryotic translation initiation factor 1A X-linked; FA, follicular adenoma; miFTC, minimally invasive FTC; wiFTC, widely invasive FTC.
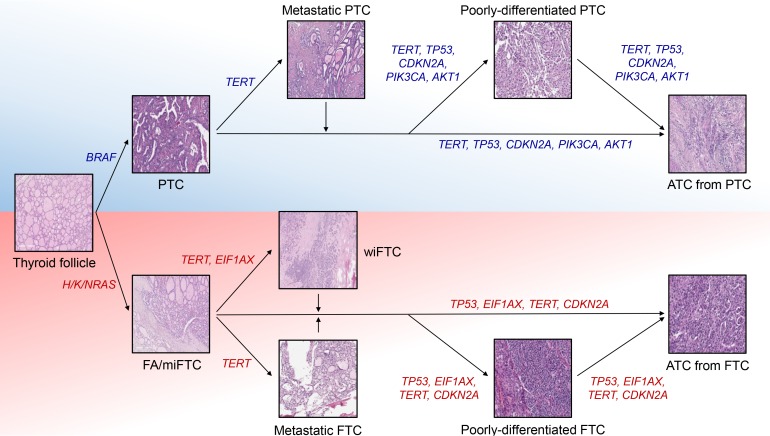
Fig. 2
Transcriptomic signatures of thyroid cancer. (A) Transcriptome based molecular subtype classifications of thyroid cancer according to histological subtypes. From The Cancer Genome Atlas (TCGA)'s original investigation, papillary thyroid cancers are classified into two molecular subtypes, BRAFV600E-like and RAS-like [10]. Afterward, Yoo et al. [11] showed that RAS-like can be breakdown into RAS-like and non-BRAF/non-RAS subtype (NBNR). RAS-like tumors with eukaryotic translation initiation factor 1A X-linked (EIF1AX), paired box 8 (PAX8)-peroxisome proliferator activated receptor gamma (PPARG), and THADA armadillo repeat containing (THADA) fusion were re-classified into NBNR. Dicer 1, ribonuclease III (DICER1), enhancer of zeste 1 polycomb repressive complex 2 subunit (EZH1), isocitrate dehydrogenase (NADP(+)) 1 (IDH1), and speckle type BTB/POZ protein (SPOP) are also associated with NBNR signature. (B) Schematic illustration of activated and deactivated signaling pathways according to the aggressiveness of thyroid cancer. BRS, BRAFV600E-RAS score; MAPK, mitogen-activated protein kinase; ECM, extracellular matrix; PD, programmed death; VEGF, vascular endothelial growth factor; JAK-STAT, Janus kinase-signal transducer and activator of transcription.
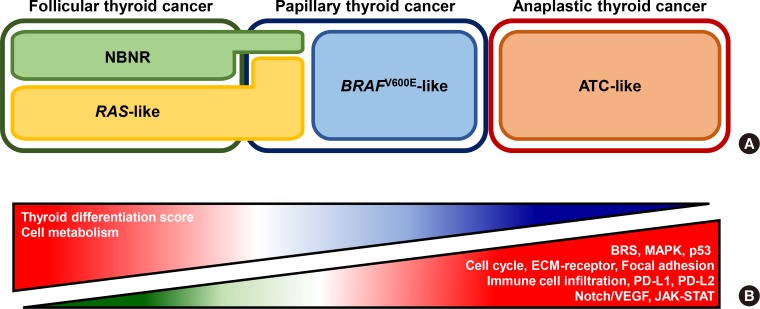
Table 1
Frequency of Commonly Altered Genes in Aggressive Thyroid Cancers
| Gene | Histological subtype | |||
|---|---|---|---|---|
| ATC | PDTC | PTC [10] | FTC | |
| BRAF | 11%–45% [151416171819202122] | 5%–33% [14151718] | 60% | 0%–8% [1113] |
| RAS | 19%–44% [141516171819202122] | 10%–38% [14151718] | 13% | 38%–50% [1113] |
| Fusion | 0%–4% [141618] | 7%–13% [1418] | 15% | 3% [11] |
| RAS+EIF1AX | 8%–30% [14151819] | 7%–11% [141518] | 0.2% | 0%–8% [1113] |
| PIK3CA | 0%–25% [1415161718] | 2%–11% [14151718] | 0.5% | 0% [1113] |
| AKT1 | 0%–8% [141516171822] | 0%–13% [14151718] | 0.8% | 0% [1113] |
| PTEN | 7%–25% [14151618] | 0%–7% [141518] | 1% | 0%–7% [1113] |
| CKDN2A | 15%–23% [141620] | 0%–7% [1418] | 0% | 0% [13] |
| TP53 | 25%–75% [141516182022] | 10%–32% [14151718] | 0.7% | 0%–3% [1113] |
| TERT | 55%–73% [1416182122] | 21%–47% [141518] | 9% | 9% [11] |
| TERT+BRAF/RAS | 25%–51% [16182122] | 5%–40% [141518] | 8% | 9% [11] |
ATC, anaplastic thyroid cancer; PDTC, poorly differentiated thyroid cancer; PTC, papillary thyroid cancer; FTC, follicular thyroid cancer; EIF1AX, eukaryotic translation initiation factor 1A X-linked; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; AKT1, AKT serine/threonine kinase 1; PTEN, phosphatase and tensin homolog; CKDN2A, cyclin dependent kinase inhibitor 2A; TP53, tumor protein p53; TERT, telomerase reverse transcriptase.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download