Abstract
References
Fig. 1
Comparison of associations between Europeans and Koreans. The P values for differentiated thyroid cancer (DTC) between Koreans (x-axis) and Europeans (y-axis) are plotted with the corresponding Korean effect sizes (odd ratio [OR]). The P value shows the −log10 scale, and the P values of novel single-nucleotide polymorphisms from this study are compared as unknown. Adapted from Son et al. [54]. FOXE1, forkhead box E1; DIRC3, disrupted in renal carcinoma 3; NKX2-1, NK2 homeobox 1; NRG1, neuregulin 1; DHX35, DEAH-box helicase 35; IMMP2L, inner mitochondrial membrane peptidase subunit 2; ARSB, arylsulfatase B; WDR11-AS1, WDR11 antisense RNA 1; VAV3, vav guanine nucleotide exchange factor 3; PCNXL2, pecanex-like 2; MSRB3, methionine sulfoxide reductase B3; SEPT11, septin 11; FHIT, fragile histidine triad; INSR, insulin receptor.
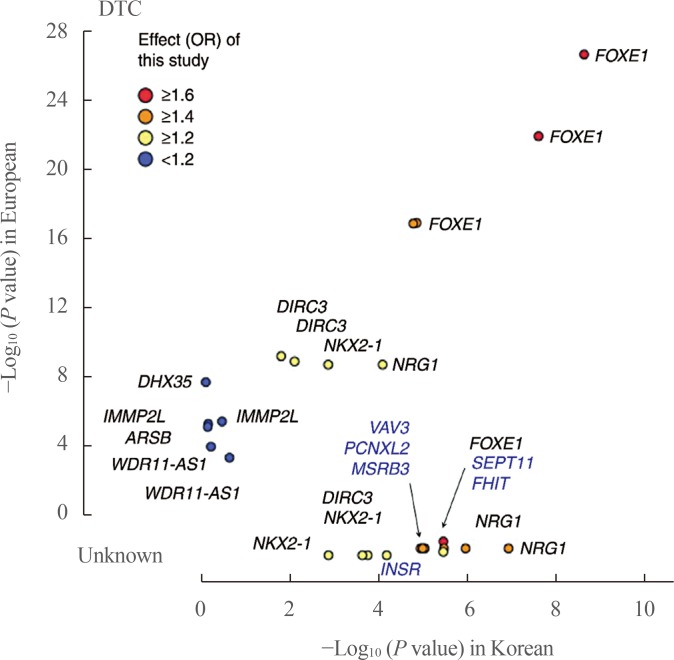
Table 1
Susceptibility Loci for Autoimmune Thyroid Disease Detected by Genome-Wide Association Studies
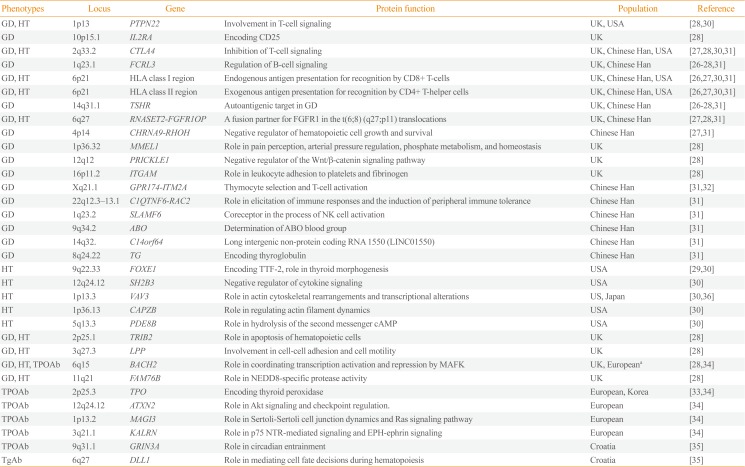
| Phenotypes | Locus | Gene | Protein function | Population | Reference |
|---|---|---|---|---|---|
| GD, HT | 1p13 | PTPN22 | Involvement in T-cell signaling | UK, USA | [2830] |
| GD | 10p15.1 | IL2RA | Encoding CD25 | UK | [28] |
| GD, HT | 2q33.2 | CTLA4 | Inhibition of T-cell signaling | UK, Chinese Han, USA | [27283031] |
| GD | 1q23.1 | FCRL3 | Regulation of B-cell signaling | UK, Chinese Han | [26272831] |
| GD, HT | 6p21 | HLA class I region | Endogenous antigen presentation for recognition by CD8+ T-cells | UK, Chinese Han, USA | [26273031] |
| GD, HT | 6p21 | HLA class II region | Exogenous antigen presentation for recognition by CD4+ T-helper cells | UK, Chinese Han, USA | [26273031] |
| GD | 14q31.1 | TSHR | Autoantigenic target in GD | UK, Chinese Han | [26272831] |
| GD, HT | 6q27 | RNASET2-FGFR1OP | A fusion partner for FGFR1 in the t(6;8) (q27;p11) translocations | UK, Chinese Han | [272831] |
| GD | 4p14 | CHRNA9-RHOH | Negative regulator of hematopoietic cell growth and survival | Chinese Han | [2731] |
| GD | 1p36.32 | MMEL1 | Role in pain perception, arterial pressure regulation, phosphate metabolism, and homeostasis | UK | [28] |
| GD | 12q12 | PRICKLE1 | Negative regulator of the Wnt/β-catenin signaling pathway | UK | [28] |
| GD | 16p11.2 | ITGAM | Role in leukocyte adhesion to platelets and fibrinogen | UK | [28] |
| GD | Xq21.1 | GPR174-ITM2A | Thymocyte selection and T-cell activation | Chinese Han | [3132] |
| GD | 22q12.3–13.1 | C1QTNF6-RAC2 | Role in elicitation of immune responses and the induction of peripheral immune tolerance | Chinese Han | [31] |
| GD | 1q23.2 | SLAMF6 | Coreceptor in the process of NK cell activation | Chinese Han | [31] |
| GD | 9q34.2 | ABO | Determination of ABO blood group | Chinese Han | [31] |
| GD | 14q32. | C14orf64 | Long intergenic non-protein coding RNA 1550 (LINC01550) | Chinese Han | [31] |
| GD | 8q24.22 | TG | Encoding thyroglobulin | Chinese Han | [31] |
| HT | 9q22.33 | FOXE1 | Encoding TTF-2, role in thyroid morphogenesis | USA | [2930] |
| HT | 12q24.12 | SH2B3 | Negative regulator of cytokine signaling | USA | [30] |
| HT | 1p13.3 | VAV3 | Role in actin cytoskeletal rearrangements and transcriptional alterations | US, Japan | [3036] |
| HT | 1p36.13 | CAPZB | Role in regulating actin filament dynamics | USA | [30] |
| HT | 5q13.3 | PDE8B | Role in hydrolysis of the second messenger cAMP | USA | [30] |
| GD, HT | 2p25.1 | TRIB2 | Role in apoptosis of hematopoietic cells | UK | [28] |
| GD, HT | 3q27.3 | LPP | Involvement in cell-cell adhesion and cell motility | UK | [28] |
| GD, HT, TPOAb | 6q15 | BACH2 | Role in coordinating transcription activation and repression by MAFK | UK, Europeana | [2834] |
| GD, HT | 11q21 | FAM76B | Role in NEDD8-specific protease activity | UK | [28] |
| TPOAb | 2p25.3 | TPO | Encoding thyroid peroxidase | European, Korea | [3334] |
| TPOAb | 12q24.12 | ATXN2 | Role in Akt signaling and checkpoint regulation. | European | [34] |
| TPOAb | 1p13.2 | MAGI3 | Role in Sertoli-Sertoli cell junction dynamics and Ras signaling pathway | European | [34] |
| TPOAb | 3q21.1 | KALRN | Role in p75 NTR-mediated signaling and EPH-ephrin signaling | European | [34] |
| TPOAb | 9q31.1 | GRIN3A | Role in circadian entrainment | Croatia | [35] |
| TgAb | 6q27 | DLL1 | Role in mediating cell fate decisions during hematopoiesis | Croatia | [35] |
GD, Graves' disease; HT, Hashimoto's thyroiditis or hypothyroidism; PTPN22, protein tyrosine phosphatase, non-receptor type 22; IL2RA, interleukin 2 receptor subunit alpha; CTLA4, cytotoxic T-lymphocyte associated protein 4; FCRL3, Fc receptor like 3; HLA, human leukocyte antigen; TSHR, thyroid stimulating hormone receptor; RNASET2, ribonuclease T2; FGFR1OP, FGFR1 oncogene partner; FGFR1, fibroblast growth factor receptor 1; CHRNA9, cholinergic receptor nicotinic alpha 9 subunit; RHOH, ras homolog family member H; MMEL1, membrane metalloendopeptidase like 1; PRICKLE1, prickle planar cell polarity protein 1; ITGAM, integrin subunit alpha M; GPR174, G protein-coupled receptor 174; ITM2A, integral membrane protein 2A; C1QTNF6, C1q and TNF related 6; RAC2, Rac family small GTPase 2; SLAMF6, SLAM family member 6; NK, natural killer; TG, anti-thyroglobulin; FOXE1, forkhead box E1; TTF-2, thyroid transcription factor-2; SH2B3, SH2B adaptor protein 3; VAV3, vav guanine nucleotide exchange factor 3; CAPZB, capping actin protein of muscle Z-line subunit beta; PDE8B, phosphodiesterase 8B; cAMP, cyclic adenosine monophosphate; TRIB2, tribbles pseudokinase 2; LPP, LIM domain containing preferred translocation partner in lipoma; TPOAb, anti-thyroid peroxidase antibody; BACH2, BTB domain and CNC homolog 2; MAFK, MAF bZIP transcription factor K; FAM76B, family with sequence similarity 76 member B; NEDD8, neural precursor cell expressed, developmentally down-regulated 8; TPO, anti-thyroid peroxidase; ATXN2, ataxin 2; MAGI3, membrane associated guanylate kinase, WW and PDZ domain containing 3; KALRN, kalirin RhoGEF kinase; NTR, neurotrophin receptor; GRIN3A, glutamate ionotropic receptor NMDA type subunit 3A; TgAb, anti-thyroglobulin antibody; DLL1, delta like canonical Notch ligand 1.
aEuropean refers to European ancestry from various countries.
Table 2
Susceptibility Loci for Levels of Thyroid Stimulating Hormone or Free Thyroxine Detected by Genome-Wide Association Studies
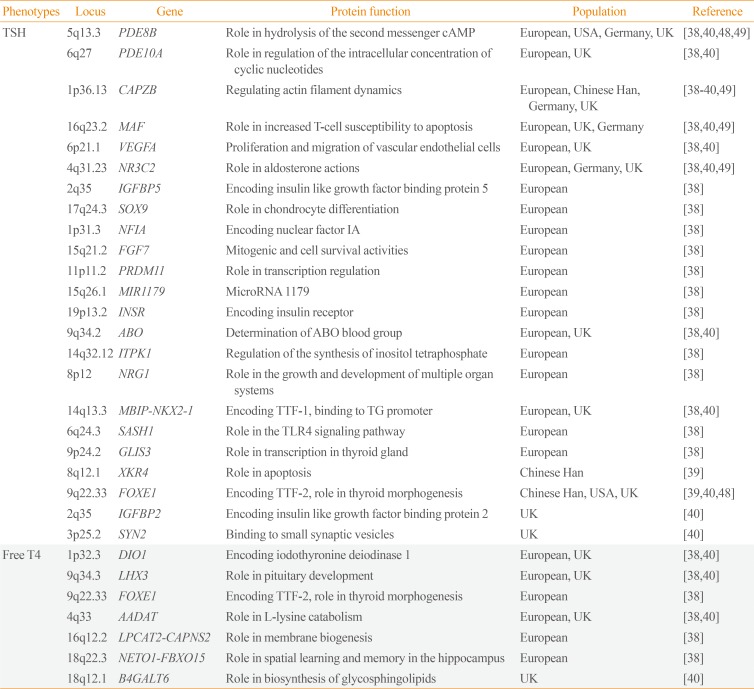
| Phenotypes | Locus | Gene | Protein function | Population | Reference |
|---|---|---|---|---|---|
| TSH | 5q13.3 | PDE8B | Role in hydrolysis of the second messenger cAMP | European, USA, Germany, UK | [38404849] |
| 6q27 | PDE10A | Role in regulation of the intracellular concentration of cyclic nucleotides | European, UK | [3840] | |
| 1p36.13 | CAPZB | Regulating actin filament dynamics | European, Chinese Han, Germany, UK | [38394049] | |
| 16q23.2 | MAF | Role in increased T-cell susceptibility to apoptosis | European, UK, Germany | [384049] | |
| 6p21.1 | VEGFA | Proliferation and migration of vascular endothelial cells | European, UK | [3840] | |
| 4q31.23 | NR3C2 | Role in aldosterone actions | European, Germany, UK | [384049] | |
| 2q35 | IGFBP5 | Encoding insulin like growth factor binding protein 5 | European | [38] | |
| 17q24.3 | SOX9 | Role in chondrocyte differentiation | European | [38] | |
| 1p31.3 | NFIA | Encoding nuclear factor IA | European | [38] | |
| 15q21.2 | FGF7 | Mitogenic and cell survival activities | European | [38] | |
| 11p11.2 | PRDM11 | Role in transcription regulation | European | [38] | |
| 15q26.1 | MIR1179 | MicroRNA 1179 | European | [38] | |
| 19p13.2 | INSR | Encoding insulin receptor | European | [38] | |
| 9q34.2 | ABO | Determination of ABO blood group | European, UK | [3840] | |
| 14q32.12 | ITPK1 | Regulation of the synthesis of inositol tetraphosphate | European | [38] | |
| 8p12 | NRG1 | Role in the growth and development of multiple organ systems | European | [38] | |
| 14q13.3 | MBIP-NKX2-1 | Encoding TTF-1, binding to TG promoter | European, UK | [3840] | |
| 6q24.3 | SASH1 | Role in the TLR4 signaling pathway | European | [38] | |
| 9p24.2 | GLIS3 | Role in transcription in thyroid gland | European | [38] | |
| 8q12.1 | XKR4 | Role in apoptosis | Chinese Han | [39] | |
| 9q22.33 | FOXE1 | Encoding TTF-2, role in thyroid morphogenesis | Chinese Han, USA, UK | [394048] | |
| 2q35 | IGFBP2 | Encoding insulin like growth factor binding protein 2 | UK | [40] | |
| 3p25.2 | SYN2 | Binding to small synaptic vesicles | UK | [40] | |
| Free T4 | 1p32.3 | DIO1 | Encoding iodothyronine deiodinase 1 | European, UK | [3840] |
| 9q34.3 | LHX3 | Role in pituitary development | European, UK | [3840] | |
| 9q22.33 | FOXE1 | Encoding TTF-2, role in thyroid morphogenesis | European | [38] | |
| 4q33 | AADAT | Role in L-lysine catabolism | European, UK | [3840] | |
| 16q12.2 | LPCAT2-CAPNS2 | Role in membrane biogenesis | European | [38] | |
| 18q22.3 | NETO1-FBXO15 | Role in spatial learning and memory in the hippocampus | European | [38] | |
| 18q12.1 | B4GALT6 | Role in biosynthesis of glycosphingolipids | UK | [40] |
TSH, thyroid stimulating hormone; PDE8B, phosphodiesterase 8B; cAMP, cyclic adenosine monophosphate; PDE10A, phosphodiesterase 10A; CAPZB, capping actin protein of muscle Z-line subunit beta; VEGFA, vascular endothelial growth factor A; NR3C2, nuclear receptor subfamily 3 group C member 2; IGFBP5, insulin like growth factor binding protein 5; SOX9, SRY-box 9; NFIA, nuclear factor I A; FGF7, fibroblast growth factor 7; PRDM11, PR/SET domain 11; MIR1179, microRNA 1179; INSR, insulin receptor; ITPK1, inositol-tetrakisphosphate 1-kinase; NRG1, neuregulin 1; MBIP, MAP3K12 binding inhibitory protein 1; NKX2-1, NK2 homeobox 1; TTF, thyroid transcription factor; TG, thyroglobulin; SASH1, SAM and SH3 domain containing 1; TLR4, Toll-like receptor 4; GLIS3, GLIS family zinc finger 3; XKR4, XK related 4; FOXE1, forkhead box E1; IGFBP2, insulin like growth factor binding protein 2; SYN2, synapsin II; T4, thyroxine; DIO1, iodothyronine deiodinase 1; LHX3, LIM homeobox 3; AADAT, aminoadipate aminotransferase; LPCAT2, lysophosphatidylcholine acyltransferase 2; CAPNS2, calpain small subunit 2; NETO1, neuropilin and tolloid like 1; FBXO15, F-box protein 15; B4GALT6, beta-1,4-galactosyltransferase 6.
Table 3
Susceptibility Loci for Thyroid Cancer Detected by Genome-Wide Association Studies
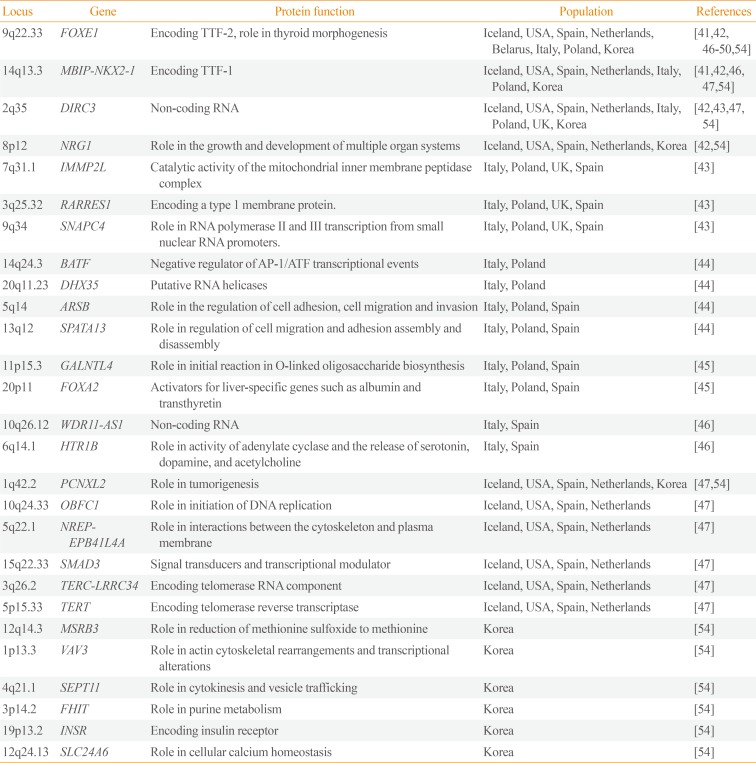
| Locus | Gene | Protein function | Population | References |
|---|---|---|---|---|
| 9q22.33 | FOXE1 | Encoding TTF-2, role in thyroid morphogenesis | Iceland, USA, Spain, Netherlands, Belarus, Italy, Poland, Korea | [4142464748495054] |
| 14q13.3 | MBIP-NKX2-1 | Encoding TTF-1 | Iceland, USA, Spain, Netherlands, Italy, Poland, Korea | [4142464754] |
| 2q35 | DIRC3 | Non-coding RNA | Iceland, USA, Spain, Netherlands, Italy, Poland, UK, Korea | [42434754] |
| 8p12 | NRG1 | Role in the growth and development of multiple organ systems | Iceland, USA, Spain, Netherlands, Korea | [4254] |
| 7q31.1 | IMMP2L | Catalytic activity of the mitochondrial inner membrane peptidase complex | Italy, Poland, UK, Spain | [43] |
| 3q25.32 | RARRES1 | Encoding a type 1 membrane protein. | Italy, Poland, UK, Spain | [43] |
| 9q34 | SNAPC4 | Role in RNA polymerase II and III transcription from small nuclear RNA promoters. | Italy, Poland, UK, Spain | [43] |
| 14q24.3 | BATF | Negative regulator of AP-1/ATF transcriptional events | Italy, Poland | [44] |
| 20q11.23 | DHX35 | Putative RNA helicases | Italy, Poland | [44] |
| 5q14 | ARSB | Role in the regulation of cell adhesion, cell migration and invasion | Italy, Poland, Spain | [44] |
| 13q12 | SPATA13 | Role in regulation of cell migration and adhesion assembly and disassembly | Italy, Poland, Spain | [44] |
| 11p15.3 | GALNTL4 | Role in initial reaction in O-linked oligosaccharide biosynthesis | Italy, Poland, Spain | [45] |
| 20p11 | FOXA2 | Activators for liver-specific genes such as albumin and transthyretin | Italy, Poland, Spain | [45] |
| 10q26.12 | WDR11-AS1 | Non-coding RNA | Italy, Spain | [46] |
| 6q14.1 | HTR1B | Role in activity of adenylate cyclase and the release of serotonin, dopamine, and acetylcholine | Italy, Spain | [46] |
| 1q42.2 | PCNXL2 | Role in tumorigenesis | Iceland, USA, Spain, Netherlands, Korea | [4754] |
| 10q24.33 | OBFC1 | Role in initiation of DNA replication | Iceland, USA, Spain, Netherlands | [47] |
| 5q22.1 | NREP-EPB41L4A | Role in interactions between the cytoskeleton and plasma membrane | Iceland, USA, Spain, Netherlands | [47] |
| 15q22.33 | SMAD3 | Signal transducers and transcriptional modulator | Iceland, USA, Spain, Netherlands | [47] |
| 3q26.2 | TERC-LRRC34 | Encoding telomerase RNA component | Iceland, USA, Spain, Netherlands | [47] |
| 5p15.33 | TERT | Encoding telomerase reverse transcriptase | Iceland, USA, Spain, Netherlands | [47] |
| 12q14.3 | MSRB3 | Role in reduction of methionine sulfoxide to methionine | Korea | [54] |
| 1p13.3 | VAV3 | Role in actin cytoskeletal rearrangements and transcriptional alterations | Korea | [54] |
| 4q21.1 | SEPT11 | Role in cytokinesis and vesicle trafficking | Korea | [54] |
| 3p14.2 | FHIT | Role in purine metabolism | Korea | [54] |
| 19p13.2 | INSR | Encoding insulin receptor | Korea | [54] |
| 12q24.13 | SLC24A6 | Role in cellular calcium homeostasis | Korea | [54] |
FOXE1, forkhead box E1; TTF, thyroid transcription factor; MBIP, MAP3K12 binding inhibitory protein 1; NKX2-1, NK2 homeobox 1; DIRC3, disrupted in renal carcinoma 3; NRG1, neuregulin 1; IMMP2L, inner mitochondrial membrane peptidase subunit 2; RARRES1, retinoic acid receptor responder 1; SNAPC4, small nuclear RNA activating complex polypeptide 4; BATF, basic leucine zipper ATF-like transcription factor; AP-1, activator protein 1; ATF, activating transcription factor; DHX35, DEAH-box helicase 35; ARSB, arylsulfatase B; SPATA13, spermatogenesis associated 13; GALNTL4, UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4; FOXA2, forkhead box A2; WDR11-AS1, WDR11 antisense RNA 1; HTR1B, 5-hydroxytryptamine receptor 1B; PCNXL2, pecanex-like 2; OBFC1, oligosaccharide-binding folds containing 1; NREP, neuronal regeneration related protein; EPB41L4A, erythrocyte membrane protein band 4.1 like 4A; SMAD3, SMAD family member 3; TERC, telomerase RNA component; LRRC34, leucine rich repeat containing 34; TERT, telomerase reverse transcriptase; MSRB3, methionine sulfoxide reductase B3; VAV3, vav guanine nucleotide exchange factor 3; SEPT11, septin 11; FHIT, fragile histidine triad; INSR, insulin receptor; SLC24A6, solute carrier family 24 member A6.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download