Abstract
Purpose
KAI1 COOH-terminal interacting tetraspanin (KITENIN) has been found to act as a promoter of metastasis in murine models of colon cancer and squamous cell carcinoma (SCC). The suppression of tumor progression and metastasis of established colon cancer in mice was observed after intravenous delivery of small interfering RNA (siRNA) targeting KITENIN. The purpose of this study was to investigate the efficacy of gene therapy targeting KITENIN in human head and neck SCC.
Materials and Methods
SNU-1041, a well-established human hypopharyngeal SCC cell line, was used. KITENIN expression in SNU-1041 was measured by Western blot analysis. The cells were prepared, maintained in culture dishes with media, and divided into two groups: the si-KITENIN group and the scrambled group (control). The siRNA targeting KITENIN (si-KITENIN) and scrambled DNA were transfected into the SNU-1041 cells in each group. The effect of gene therapy was compared by in vitro experiments to evaluate invasion, migration, and proliferation.
Results
KITENIN was strongly expressed in the SNU-1041 cells, and the number of invaded cells was reduced more in the si-KITENIN group than in the scrambled group (p<0.001). The speed for the narrowing gap, made through adherent cells, was lower in the si-KITENIN group (p<0.001), and the number of viable proliferating cells was reduced in the si-KITENIN group compared to the scrambled group (p<0.001, the third day). KITENIN protein expression was no longer identified in the si-KITENIN group.
Reducing the progression and metastasis of head and neck squamous cell carcinoma (SCC) requires an understanding of molecular mechanisms involved in invasion and dissemination.1 Gene therapy for tumor metastasis is a promising because it inhibits the spread of cancer regardless of the tumorigenicity.2,3
A large body of evidence suggests that down-regulation of KAI1/CD82 expression occurs with invasive and metastatic disease, and that it is an important step in the progression of many human malignancies.4-8 KAI1, a tumor metastasis suppressor gene, is a transmembrane glycoprotein that is a member of the tetraspanin superfamily. The metastasis suppressor function was recently reported to be decreased in a spliced variant of KAI1 at the COOH-terminal region, suggesting that the COOH-terminal region of KAI1 is important for the effects of KAI1 on cell motility.9 Subsequently, a protein that interacted with the COOH-terminal cytoplasm domain of KAI1 was identified via a two-hybrid yeast system.10 The function of this protein Vang-like 1 (VANGL111) during carcinogenesis has not been fully described, and it has been renamed as KAI1 COOH-terminal interacting tetraspanin (KITENIN).
KITENIN-overexpressing CT-26 mouse colon cancer cells have shown increased tumorigenicity and early hepatic metastasis in vivo, as well as increased invasiveness and adhesiveness to fibronectin in vitro, compared to parental cells.10 Moreover, the suppression of progression and metastasis of established colon cancer has been observed in mice after intravenous delivery of small interfering RNA (siRNA) targeting KITENIN.12
We recently reported that KITENIN promoted pulmonary metastasis in a murine model of SCC by increasing cancer cell invasion and adhesion to fibronectin,13 and that KITENIN represented a more aggressive phenotype in a murine model of oral cavity squamous carcinoma.14 Furthermore, we investigated the expression of KITENIN in human laryngeal SCC, and found that KITENIN expression was significantly increased in laryngeal cancer tissues, compared to the adjacent normal tissue mucosa, as well as in metastatic lymph nodes compared to non-metastatic lymph nodes. High KITENIN expression was associated significantly with an advanced disease stage, extent of the tumor, and lymph node metastasis.15
siRNA is a small double-stranded, non-protein coding RNA (21-31 nucleotides) involved in gene silencing functions, especially RNA interference (RNAi). This ability of siRNA has provided researchers with a novel tool to block the expression of disease-causing genes, provided that their mRNA sequences are known. siRNAs can be delivered to cells either exogenously as synthetic agents or endogenously as gene-coding siRNAs. Recent studies have demonstrated the general application of siRNAs to silence gene expression in a range of cell types and in mammalian models.16
The purpose of this study was to investigate the efficacy of gene therapy (siRNA targeting KITENIN) in human head and neck SCC.
SNU-1041 cells,17-19 a well-established human head and neck SCC cell line, were grown in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA) in a humidified atmosphere of 5% CO2 at 37℃. The cells were prepared and maintained in culture dishes with media, and divided into two groups: the si-KITENIN group and the scrambled group (control). The siRNA targeting KITENIN (si-KITENIN) and the scrambled DNA were transfected into the SNU-1041 cells in each group. The sequence of siRNA was as follows: human KITENIN 5'-GCUUGGACUUCAGCCUCGUAGUCAA-3'. The transfections were performed using the Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. The cells were maintained and used for further analysis.
Western blot analysis was used to investigate KITENIN expression before and after transfection of siRNA (si-KITENIN). The tissues were solubilized in NP40 lysis buffer containing a protease inhibitor mixture (Roche, Indianapolis, IN) and 1 mM phenylmethylsulfonyl fluoride. The resolved proteins (50 µg) were transferred to a nitrocellulose membrane and blotted with the KITENIN antibody and antirabbit immunoglobulin-horseradish peroxidase (Amersham, Arlington Heights, IL, USA). The blot was reprobed with anti-actin antibody (I-19; Santa Cruz Biotechnology, Santa Cruz, CA, USA) to control loading variations.
Cell migration was measured using a Transwell migration apparatus (Costar Inc., Cambridge, UK) with minor modification as described previously.9 The filters (8-µm pore size) were coated with 1% gelatin solution on both the top and bottom surfaces. The cells from each group (si-KITENIN group, scrambled group) were harvested, washed once in serum-free DMEM/0.2% bovine serum albumin (BSA), and resuspended at 2×106 cells/µL DMEM/0.2% BSA. To start the assay, 120 µL (2.4×105 cells) was loaded onto the upper chamber of the Transwell, whereas 400 µL DMEM/0.2% BSA containing 20 µg/mL human plasma fibronectin (Calbiochem, La Jolla, CA, USA), a chemotactic factor, was loaded onto the lower chamber. The Transwell apparatus was incubated for 24 hours at 37℃. At the end of incubation, the cells attached to the membranes were fixed and stained with Diff-Quick (International Reagents, Kobe, Japan) by following the manufacturer's protocol. The cells on the top surface of the filters were wiped off with cotton balls, and the cells that migrated to the bottom surface were counted in ten random squares of 0.5×0.5 mm2 by microscopic field of view. The results were expressed as mean±SE of the number of cells/field.
si-KITENIN-transfected and scrambled DNA-transfected SNU-1041 cells were seeded in a 6-well plate (1×105 cells/well). Twenty-four hours later, the media were changed to serum-free DMEM and incubated for 12 hours. After the media were removed from the wells, a straight transverse line through the adherent cells was drawn using a ruler and a 1000 µL-tip, resulting in a uniform gap. The media were changed to DMEM supplemented with 2% fetal bovine serum. At 0, 20, 28, and 44 hours later; the distances between the gaps were measured in centimeters after capture of six random sites by the microscopic field of view.
The proliferation and viability of the cells were measured using an enhanced cell viability assay kit, EZ CyTox (Daeil Lab Service Co., Seoul, Korea). The cells were seeded at 2×104 cells/well in 48-well plates, transfected with si-KITENIN and scrambled DNA, and grown overnight in the presence of serum. Next, EZ CyTox was added according to the manufacturer's instructions for 2 hours, after which the discolored cells were moved into 96-well plates for analysis. Absorbance at 450 nm was determined using a microplate reader with SOFTmax PRO software (Molecular Devices, Sunnyvale, CA, USA).
Fig. 1 shows that KITENIN was strongly expressed in the SNU-1041 cells and scrambled DNA-transfected SNU-1041 cells (SNU-1041/Scrambled). However, the expression of KITENIN protein was not observed in the si-KITENIN-transfected SNU-1041 cells (SNU-1041/si-KITENIN), demonstrating that KITENIN was effectively blocked by siRNA.
As shown in Fig. 2, the number of invading si-KITENIN-transfected SNU-1041 cells was 64.2±19.2, whereas the number of the scrambled DNA-transfected SNU-1041 cells was 98.6±10.2, as measured by 10 random squares of 0.5×0.5 mm2 by the microscopic field of view under conditions with 20 µg/mL of fibronectin; the difference between the two groups was statistically significant (p<0.001).
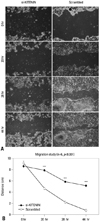
The artificial wound gap became significantly narrower in the control group as time passed by 20, 28, 44 hours, compared to the si-KITENIN group (p<0.001)(Fig. 3). For the scrambled group, the gap was nearly filled within 44 hours, however, it remained wide open in the si-KITENIN group.
The number of proliferating cells, as determined by absorbance, was significantly decreased in the si-KITENIN group compared to the scrambled group (Fig. 4). On the third day, absorbance was 0.4933±0.0484 in the si-KITENIN group, whereas 0.6023±0.0319 in the scrambled group (p<0.001).
siRNA, known as RNAi, is a powerful tool for strong and specific suppression of gene expression.16 The use of these molecules can be applied to a wide range of cancers and other proliferative disorders where aberrant gene expression occurs. Therefore, oncogenic and mutant tumor suppressor genes might represent potential targets for the RNAi approach. It has been shown that mutated p53 protein, involved in almost half of human malignancies, was eliminated by siRNA and the function of wild-type p53 restored.20 Other examples include: elimination of the Ras oncogene using siRNAs,21 transfection of leukemic cells with siRNAs targeting a BCR-ABL fusion transcript, thereby inducing apoptosis,22,23 and targeting of VEGF by siRNAs.24
In this study, siRNA targeting KITENIN was effective. SNU-1041, an established human head and neck squamous carcinoma cell line,17-19 expressed very high levels of KITENIN, which functions as a metastasis-promoting gene. Our previous data demonstrated that many human head and neck squamous cancer tissues expressed KITENIN, and that the level of KITENIN correlated with the disease stage. After transfection of si-KITENIN into SNU-1041 cells, KITENIN was no longer identified in the cells. siRNA-transfected SNU-1041 cells showed reduced in vitro invasion, migration, and proliferative characteristics compared to the cells in the control group.
siRNAs can be delivered in two ways: exogenous administration of synthetic siRNAs and endogenous expression of siRNA via plasmid or viral vectors.
Generally, synthetic siRNAs are delivered to cells in culture via liposome-based transfection reagents. In this study, we used the lipofectamine trasfection reagent (RNAiMAX, Invitrogen) and the experiments were successful. However, this delivery offers only immediate and/or short-term effects.25 Therefore, siRNA delivery systems which can provide long-term biological effects are needed.
To overcome these problems, several research groups have shown that short hairpin siRNA can be produced from expression plasmids that contain promoters that are dependent on either RNA polymerase (pol) II or pol III.26-29 Brummelkamp, et al.26 developed a new expression vector system, pSUPER (suppression of endogenous RNA) that directs the synthesis of siRNA-like transcripts in mammalian cells, and Lee, et al.12 used the pSUPER vector system to deliver KITENIN siRNA for efficient and stable suppression of KITENIN expression in established tumors of syngeneic mice. The latter observed marked inhibition of the growth of established tumors as well as suppression of distant metastases of colon cancer, following four weekly or semiweekly intravenous injections of pSUPER-KITENIN. In addition to plasmid vectors, adenoviral and lentiviral-based vectors have also been developed and used successfully.30,31 For our next in vivo study with animals on anti-KITENIN delivery, plasmid or viral vector systems will be studied.
In conclusion, gene therapy using anti-KITENIN strategies may help or delay the progression (invasion, migration and proliferation) of head and neck squamous carcinoma.
Figures and Tables
Fig. 1
KITENIN expression in SNU-1041 cells: KITENIN was strongly expressed in SNU-1041 cells and scrambled DNA-transfected SNU-1041 cells (SNU-1041/Scrambled). The expression of KITENIN protein was not observed in si-KITENIN-transfected SNU-1041 cells (SNU-1041/si-KITENIN).
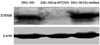
Fig. 2
Cell invasion assay: the number of invading si-KITENIN-transfected SNU-1041 cells (A) was 64.2±19.2, whereas the number was 98.6±10.2 for the scrambled DNA-transfected SNU-1041 cells (B), as measured by 10 random squares of 0.5×0.5 mm2 on the microscopic field of view under conditions with 20 µg/mL of fibronectin; the difference between the two groups was statistically significant (p<0.001)(C).
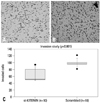
Fig. 3
Cell migration assay: the artificial wound gap became significantly narrower in the control group as time passed by 20, 28, 44 hours, compared to the si-KITENIN group (p<0.001)(B). In the scrambled group, the gap was nearly filled within 44 hours. However, it remained wide open in the si-KITENIN group (A).
ACKNOWLEDGEMENTS
This work was supported by the Korean Research Foundation grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2008-1684).
References
1. Howell GM, Grandis JR. Molecular mediators of metastasis in head and neck squamous cell carcinoma. Head Neck. 2005. 27:710–717.

2. Woodhouse EC, Chuaqui RF, Liotta LA. General mechanisms of metastasis. Cancer. 1997. 80:1529–1537.

3. Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr Opin Cell Biol. 2005. 17:559–564.

4. Jackson P, Marreiros A, Russell PJ. KAI1 tetraspanin and metastasis suppressor. Int J Biochem Cell Biol. 2005. 37:530–534.

5. Takaoka A, Hinoda Y, Satoh S, Adachi Y, Itoh F, Adachi M, et al. Suppression of invasive properties of colon cancer cells by a metastasis suppressor KAI1 gene. Oncogene. 1998. 16:1443–1453.

6. Yang JL, Jackson P, Yu Y, Yoshie O, Ham JM, Russell PJ, et al. Invasion and metastasis correlate with down-regulation of KAI1 expression in human colon cancer cell lines. GI Cancer. 2001. 3:313–322.
7. Yang X, Wei LL, Tang C, Slack R, Mueller S, Lippman ME. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 2001. 61:5284–5288.
8. Jee B, Jin K, Hahn JH, Song HG, Lee H. Metastasis-suppressor KAI1/CD82 induces homotypic aggregation of human prostate cancer cells through Src-dependent pathway. Exp Mol Med. 2003. 35:30–37.

9. Lee JH, Seo YW, Park SR, Kim YJ, Kim KK. Expression of a splice variant of KAI1, a tumor metastasis suppressor gene, influences tumor invasion and progression. Cancer Res. 2003. 63:7247–7255.
10. Lee JH, Park SR, Chay KO, Seo YW, Kook H, Ahn KY, et al. KAI1 COOH-terminal interacting tetraspanin (KITENIN), a member of the tetraspanin family, interacts with KAI1, a tumor metastasis suppressor, and enhances metastasis of cancer. Cancer Res. 2004. 64:4235–4243.

11. Yagyu R, Hamamoto R, Furukawa Y, Okabe H, Yamamura T, Nakamura Y. Isolation and characterization of a novel human gene, VANGL1, as a therapeutic target for hepatocellular carcinoma. Int J Oncol. 2002. 20:1173–1178.

12. Lee JH, Cho ES, Kim MY, Seo YW, Kho DH, Chung IJ, et al. Suppression of progression and metastasis of established colon tumors in mice by intravenous delivery of short interfering RNA targeting KITENIN, a metastasis-enhancing protein. Cancer Res. 2005. 65:8993–9003.

13. Lee JK, Bae JA, Sun EG, Kim HD, Yoon TM, Kim K, et al. KITENIN increases invasion and migration of mouse squamous cancer cells and promotes pulmonary metastasis in a mouse squamous tumor model. FEBS Lett. 2009. 583:711–717.

14. Lee JK, Lim SC, Kim HD, Yoon TM, Kim K, Nam JH, et al. KITENIN represents a more aggressive phenotype in a murine model of oral cavity squamous carcinoma. Otolaryngol Head Neck Surg. 2010. 142:747–752.

15. Lee JK, Yoon TM, Seo DJ, Sun EG, Bae JA, Lim SC, et al. KAI1 COOH-terminal interacting tetraspanin (KITENIN) expression in early and advanced laryngeal cancer. Laryngoscope. 2010. 120:953–958.

17. Kim SG, Hong JW, Boo SH, Kim MG, Lee KD, Ahn JC, et al. Combination treatment of Cetuximab and photodynamic therapy in SNU-1041 squamous cancer cell line. Oncol Rep. 2009. 22:701–708.

18. Koh TY, Park SW, Park KH, Lee SG, Seol JG, Lee DW, et al. Inhibitory effect of p27KIP1 gene transfer on head and neck squamous cell carcinoma cell lines. Head Neck. 2003. 25:44–49.

19. Sung MW, Roh JL, Park BJ, Park SW, Kwon TK, Lee SJ, et al. Bile acid induces cyclo-oxygenase-2 expression in cultured human pharyngeal cells: a possible mechanism of carcinogenesis in the upper aerodigestive tract by laryngopharyngeal reflux. Laryngoscope. 2003. 113:1059–1063.

20. Martinez LA, Naguibneva I, Lehrmann H, Vervisch A, Tchénio T, Lozano G, et al. Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc Natl Acad Sci U S A. 2002. 99:14849–14854.

21. Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002. 2:243–247.

22. Scherr M, Battmer K, Winkler T, Heidenreich O, Ganser A, Eder M. Specific inhibition of bcr-abl gene expression by small interfering RNA. Blood. 2003. 101:1566–1569.

23. Wilda M, Fuchs U, Wössmann W, Borkhardt A. Killing of leukemic cells with a BCR/ABL fusion gene by RNA interference (RNAi). Oncogene. 2002. 21:5716–5724.

24. Zhang L, Yang N, Mohamed-Hadley A, Rubin SC, Coukos G. Vector-based RNAi, a novel tool for isoform-specific knock-down of VEGF and anti-angiogenesis gene therapy of cancer. Biochem Biophys Res Commun. 2003. 303:1169–1178.

25. Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002. 30:1757–1766.

26. Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002. 296:550–553.

27. Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002. 20:497–500.

28. Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002. 20:500–505.

29. Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002. 20:1006–1010.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download