Lung cancer remains the leading cause of cancer-related deaths in Korea and worldwide.
12 The major method of screening, low-dose computed tomography (LDCT), was validated in the landmark National Lung Screening Trial (NLST), where LDCT demonstrated a 20% relative reduction in lung cancer-related mortality in a high-risk population.
3
Although LDCT screening for lung cancer is associated with the low specificity (73.4%)
4 and the resulting high rate of false-positives (96.4%),
3 annual LDCT-screening for lung cancer was recently recommended for high-risk individuals.
5 However, in this recommendation, the need for more research into the use of biomarkers to complement LDCT screening was also stressed.
56
Aptamers, defined as single chain nucleotide ligands, have been developed as novel capture arrays that share many features with antibodies.
78 Owing to its innate property,
8 aptamer technology enables the development of a highly multiplexed proteomic assay to analyze multiple proteins in complex biological samples.
9 Recently, an aptamer-based high-throughput proteomics assay, the slow off-rate modified aptamers (SOMAmers) scan was developed
10 and it identified several candidate biomarkers for non-small cell lung cancers (NSCLCs).
11 Mehan and colleagues
12 discovered that 15 robust biomarkers was selected using the SOMA scan and a 7-marker random forest classifier was built. These studies have supported the application of aptamers in the development of lung cancer biomarkers in the US population with the potential to complement the current screening standards. Our previous study investigated candidate biomarkers in the Korean population using a new modified aptamer-based proteomic technology and developed a 7-protein panel (AptoDetect™-Lung, Aptamer Sciences Inc., Pohang, Korea) that discriminates lung cancers from controls with an area under the curve (AUC) of 0.82 and 0.77 in the training and verification sets, respectively.
13
In this study, we sought to validate the performance of AptoDetect™-Lung for lung cancer detection in an independent and larger Korean population.
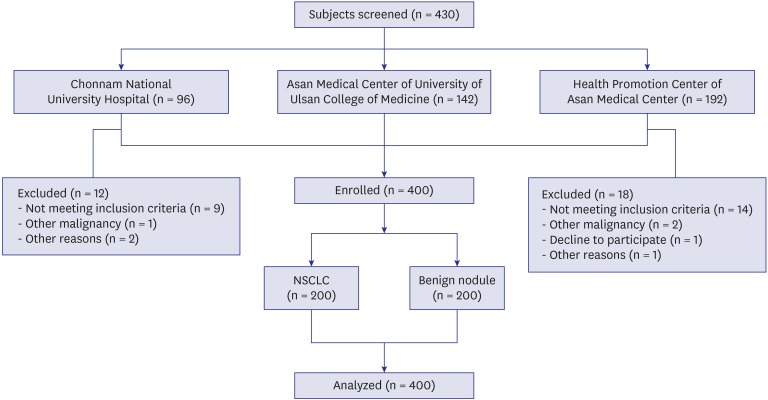
A total of 430 participants were screened, and 200 patients with biopsy-proven NSCLC and 200 participants as benign nodule controls were enrolled from three study centers; the two lung cancer cohorts of the Chonnam National University Hospital, the Asan Medical Center of University of Ulsan College of Medicine, and a Health Promotion Center of the Asan Medical Center (
Fig. 1).
The subject exclusion criteria were age < 20 or > 80 years, the lack of nodule size or histopathologic cancer diagnosis, a diagnosis of small-cell lung cancer or a history of other malignancy.
Clinical data and blood samples were collected from the NSCLC group diagnosed with pathologic or clinical stage I–IV NSCLC. Blood samples were collected within 4 weeks of the first biopsy-proven lung cancer diagnosis and before treatment or removal of the tumor by standard surgical procedures. The benign nodule control group consisted of subjects who received a lung health screening exam from November 2016 to February 2017. Chest computed tomography (CT) and blood sampling was collected simultaneously with the CT scan. All participants were followed with LDCT and determined to be cancer-free after a minimum of 1-year follow-up.
In order to assess the samples, biomarkers were measured using the AptoDetect™-Lung kit, which was developed in a previous study.
13 Briefly, to select of the panel of the biomarker, we used the Naïve Bayes method to systematically assess potential biomarker performance, and the Naïve Bayes classifier was developed a diagnostics model that generated the probability a patient has lung cancer given their protein biomarker levels. Serum concentrations of seven proteins (EGFR1, MMP7, CA6, KIT, CRP, C9, and SERPINA3) were measured using an aptamer-based multiplex assay and the likelihood ratio was calculated for each sample. To simplify the score, the likelihood ratio was transformed to a risk stratification score spanning 0–10. We employed a cut-off of 5 points, which had 75% sensitivity and 91.7% specificity for NSCLC patients against benign nodule controls.
13 Samples with a score ≥ 5 were considered positive and high-risk, whereas samples with a score < 5 were considered negative and low-risk. As described in a previous report, we studied the accuracy of AptoDetect™-Lung by comparing sensitivity and specificity and used the AUC of the receiver operating characteristic curve to compare candidate classifier performance.
13
Student's t-tests or Mann-Whitney U tests were used to compare continuous variables, and χ2 tests or Fisher's exact tests were used to compare categorical variables. Analyses were performed using SPSS for Windows Version 21.0 (SPSS Inc., Chicago, IL, USA) and SAS for Windows Version 9.4 (SAS Institute, Cary, NC, USA). Two-sided P values < 0.05 were considered to be statistically significant.
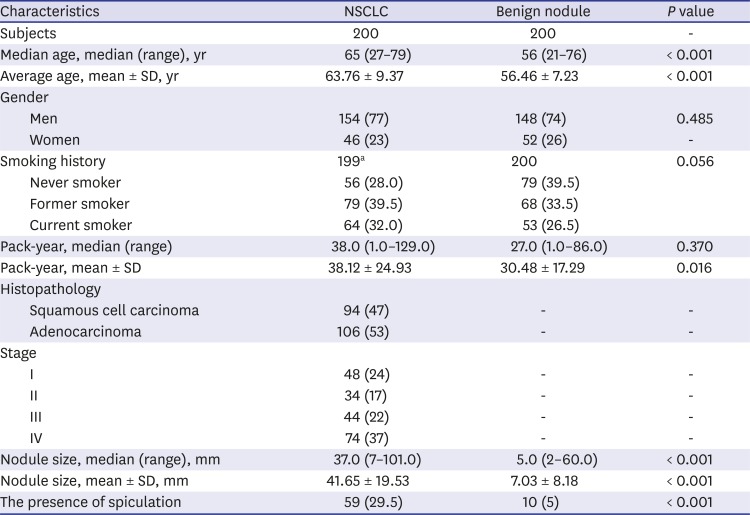
The median age of the NSCLC group was 65 years (range, 27–79 years), whereas the median age of the benign nodule control group was 56 years (range, 21–76 years). There was no difference in gender and smoking history between two groups. The mean pack-years in the NSCLC was significantly higher than that of the benign nodule control (38.12 ± 24.93 vs. 30.48 ± 17.29;
P = 0.016). In the NSCLC, 53% (106/200) of cases were adenocarcinoma and 47% (94/200) of cases were squamous cell carcinoma (
Table 1).
Based on AptoDetect™-Lung scores, the NSCLC and benign nodule control groups were divided into low-risk (negative) (< 5) and high-risk (positive) (≥ 5). Of the 171 cases categorized as high-risk, 137 (80.1%) were NSCLC cases and 34 (19.9%) were benign nodule control cases. The 229 cases categorized as low-risk included 166 (72.5%) benign nodule control and 63 (27.5%) NSCLC.
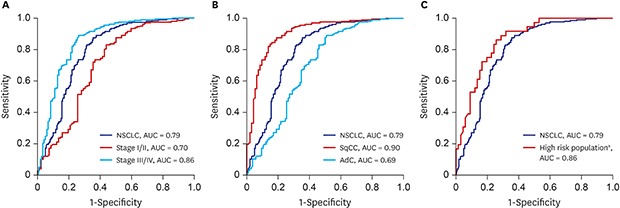
The result of clinical performance showed that AptoDetect™-Lung for discriminating NSCLC from benign nodule controls had a sensitivity and specificity of 68.5% and 83.0%, respectively, and the AUC was 0.79. The AUC of the AptoDetect™-Lung in discriminating between early stages (I and II) and benign nodule controls was 0.70, and it was 0.86 for discriminating between stage III and IV and benign nodule controls (
Fig. 2A). AptoDetect™-Lung also accurately divided the most prevalent NSCLC histological types; the AUC for squamous cell carcinoma was 0.90 and the AUC for adenocarcinoma was 0.69 (
Fig. 2B).
The high-risk population (n = 128) in this study corresponding to more than 55 years of age and a smoking history of 30 pack-years consisted of 86 (67.2%) NSCLC and 42 (33.1%) benign nodule control cases. AptoDetect™-Lung had a sensitivity and specificity of 73.3% and 90.5%, respectively, and the AUC was 0.88, indicating the good discrimination among this group (
Fig. 2C).
The main concern raised by clinicians performing lung cancer CT screening is the high prevalence of one or more lung nodules, with many being undiagnosed as either malignant or benign.
36 In Korea, LDCT screening showed that 35% of subjects had at least one or more non-calcified lung nodule, and lung cancer detection rates were 0.36%.
14 In the organized, controlled setting of LDCT randomized and cohort screening studies, the rate of invasive procedures was low, varying from 1% to 4%.
15 However, approximately 25% of invasive procedures are performed in patients with nodules that are eventually shown to be benign (range, 0%–45%).
316 Consequently, there is an unmet need for biomarkers that can discriminate between benign and malignant nodules to complement CT-based diagnosis. This corresponds to the specificity of the test in determining the percentage of benign nodules correctly called benign (that is, negative) by the test. Thus, the higher the specificity at a given high negative predictive value, the more patients with benign nodules that do not have to undergo the unnecessary invasive procedures.
17 AptoDetect™-Lung suggested more than 90% specificity in the high-risk population. This is similar to the result from the EarlyCDT
®-Lung (Innovative Diagnostic Laboratory, Richmond, VA, USA) test, which showed 90% specificity and 41% sensitivity.
18
AptoDetect™-Lung showed an excellent AUC of 0.90 for squamous cell carcinoma. NLST showed that by tumor histology, mortality risk ratio was 0.75 for adenocarcinoma, 0.71 for all NSCLC except squamous, 1.23 for squamous cell carcinoma.
19 In addition, squamous cell carcinoma is the NSCLC histology most commonly undetected by CT, perhaps because the central tumor location obscures CT detection and it tends to have a rapid growth rate that can lead to diagnosis as interval tumors.
13 Therefore, AptoDetect™-Lung has the advantage of high specificity for detecting squamous cell carcinoma, and it might have a complementary role in lung cancer screening.
Currently, Lung Imaging Reporting and Data System (Lung-RADS) which defines a positive screening test and provides patient's clinical management recommendations based on level of risk, has been adopted at many academic medical centers in the US.
20 Integration of AptoDetect™-Lung into Lung-RADS may increase the accuracy of nodule classification, and suggest the strategy to guide the lung nodule management leading to a decrease in unnecessary follow-up imaging or invasive procedures, and potentially avoid unnecessary morbidity, mortality, and health care costs.
AptoDetect™-Lung offers the best validated performance to discriminate NSCLC from benign nodules in a high-risk population that corresponds to more than 55 years of age and a smoking history of at least 30 pack-years and could serve a complementary role in lung cancer screening. Additionally, large prospective trials are needed to validate the improved efficacy and utility in clinical practice for screening and diagnostic processes by pairing AptoDetect™-Lung with clinical management recommendations in lung cancer screening CT.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download