This article has been
cited by other articles in ScienceCentral.
Abstract
Pathogenic gram-negatives that produce 16S ribosomal RNA methyltransferases (16S RMTases) have already been distributed all over the world. To investigate the predominance of aminoglycoside resistance associated with 16S RMTases in Korea, we collected a total of 222 amikacin resistant Gram-negative clinical isolates from patient specimens between 1999 and 2015 from three hospital banks across Korea. ArmA and rmtB were the predominant 16S RMTase genes responsible for aminoglycoside-resistant isolates circulating in Korean community settings although only one rmtA-producing isolate was detected in 2006.
Keywords: Aminoglycoside Resistance, ArmA, RmtB, Korea
Aminoglycosides are one of the key classes of antimicrobial agents used in the treatment of Gram-negative bacterial infections. These agents bind to the highly conserved A site of the 16S rRNA of the bacterial 30S ribosomal subunits, thereby causing interference with bacterial protein synthesis, which ultimately leads to bacterial death.
1 However, the increase in aminoglycoside-resistant Gram-negative bacteria in recent years is a multi-faceted issue requiring urgent attention. Although several pathways that provide resistance to aminoglycoside antibiotics are known, exogenously acquired 16S ribosomal RNA methyltransferases (16S RMTases) have emerged as a major mechanism of high-level resistance to most clinically important aminoglycosides, including arbekacin, amikacin, tobramycin, and gentamicin in Gram-negative pathogens.
1 In 2003, the first acquired 16S RMTase genes,
armA and
rmtA, were identified in
Klebsiella pneumoniae and
Pseudomonas aeruginosa, respectively.
2 Since then, other plasmid-mediated 16S RMTase genes (
rmtB through
rmtH, and
npmA) have been found in clinical isolates.
234 Among them,
armA and
rmtB have been found in many species of Gram-negative bacilli in Asia.
5
In a Korean nationwide surveillance, the amikacin resistance rates of
K. pneumoniae increased from 8% in 1997 to 13% in 2003.
67 The frequency of high-level resistance to amikacin or arbekacin was 9.5% (15/158), 10.3% (13/126), and 17.1% (22/129) for
Enterobacter cloacae,
Citrobacter freundii and
Serratia marcescens isolates, respectively.
8 In another study, 24 hospitals and two Community Labs participated in collecting antimicrobial susceptibility data in Korea, and the resistance rates of
K. pneumoniae,
E. cloacae,
S. marcescens,
Acinetobacter spp., and
P. aeruginosa to amikacin were high in the hospital laboratories: 15%, 5%, 10%, 48%, and 19%, respectively.
9 Taken together, these results suggest that 16S RMTase-producing bacteria are becoming prevalent in Korea and highlight the need for continued surveillance to investigate the overall trend of aminoglycoside resistance among clinically important pathogens. To provide ongoing insight into aminoglycoside resistance, in this study, we investigated predominant 16S RMTases in Korea from our data and from previously published literatures.
To examine the recent predominance of aminoglycoside resistance associated with 16S RMTases in Korea, a total of 222 potential amikacin resistant Gram-negative clinical isolates from patient specimens were collected between 1999 and 2015 from three hospital banks across Korea and primarily identified by each bank through Vitek Mass Spectrometry: 73 isolates from Chonbuk National University Hospital Culture Collection for Pathogens from 2001 to 2013; 88 isolates from Gyeongsang National University Hospital Culture Collection for Pathogens from 1999 to 2015; 61 isolates from Kyungpook National University Hospital Culture Collection for Pathogens from 2012 to 2015. The specimens were from the following sources: 10 peritoneal fluid, 77 blood, 27 open wound, 6 bile acid, 51 urine, 46 sputum and bronchial washing, 3 ear discharge, and 2 pleural fluid. To further confirm amikacin-resistance and determine the minimum inhibitory concentrations (MICs) of amikacin for the collected isolates, we evaluated the susceptibility to amikacin by using the Etest. Briefly, Etest strips (bioMérieux, Marcy-l'Étoile, France) with amikacin were placed on Mueller-Hinton (MH) agar plates lawn-inoculated with suspension of isolates grown to an optical density of 0.5 McFarland units to determine the MICs of amikacin for each isolate. After incubation, MICs were read directly from the Etest strip described previously.
10 We also screened the amikacin-resistant isolates using polymerase chain reaction (PCR) to detect the known 16S RMTase genes
armA,
rmtA,
rmtB,
rmtB2,
rmtC,
rmtD,
rmtE,
rmtF,
rmtG, and
rmtH. Briefly, DNA template for PCR were prepared from bacterial isolates cultured in LB broth containing amikacin (10 μg/mL) and previously described oligonucleotides were used to amplify gene fragments from 16S RMTases.
1112 The PCR products were sequenced, and the sequences were compared with those in the GenBank nucleotide database. A total of 198 (89.2%) isolates were confirmed as amikacin-resistant (> 16 μg/mL) among the 222 collected isolates primarily confirmed as potential aminoglycoside-resistance at the three hospitals in Korea (
Table 1). Among them, 190 (96.0%) were highly resistant to amikacin (> 256 μg/mL), a resistance phenotype consistent with production of 16S RMTase. By PCR, 142 (74.7%) of the high level-amikacin resistant bacteria were positive for 16S RMTase genes; 141
armA and one
rmtB.
To further investigate whether aminoglycoside resistance was transferable, plasmids from clinical isolates with high-level resistance to amikacin (> 256 μg/mL) and with 16S RMTase genes (armA or rmtB) were prepared using the alkaline lysis method and were electro-transformed into Escherichia coli TOP10 competent cells (Invitrogen, Carlsbad, TX, USA). Transformants were selected on LB agar plates containing 50 μg/mL gentamicin, and gentamicin-resistant transformants were tested for susceptibility to amikacin (256 μg/mL). The plasmids from transformants with both gentamicin and amikacin resistance were isolated, used as template DNAs for the PCR-based detection of 16S RMTases, and analyzed against the GenBank nucleotide database, as described above. Of six armA and one rmtB from the high-level amikacin-resistant isolates tested, all transferred resistance to amikacin through plasmids.
We reviewed 419 articles using the keywords “16S rRNA methyltransferase” or “16S methyltransferase” or “Korea” in PubMed for articles published from 1974 to 2017 to study overall trends over time of 16S RMTases from clinical isolates in Korea. A total of 10 papers related to 16S RMTases from clinical isolates were published in Korea from 2006 to 2017 (
Table 2).
781314151617181920 Among 331 amikacin resistant gram-negative organism (63
E. coli; 149
K. pneumoniae; 2
K. spp.; 8
C. freundii; 27
E. cloacae; 7
E. spp.; 14
S. marcescens; 30
Acinetobacter spp.; 27
P. aeruginosa; 3
Providencia spp.; 1
Morganella morganii),
armA was 192 and
rmtB was 52 isolates.
71415 In 145 high-level amikacin resistant isolates,
armA was 110 and
rmtB was 19 isolates.
816 In the other 5 articles,
armA was detected 30 of 44 extended spectrum β-lactamase producing
K. pneumonia, 14 of 111 multi-drug resistant (MDR)
P. aeruginosa, 28 of 31 MDR
Acinetobacter baumannii, 4 of 264 isolates from urines.
13171920 One
rmtB was from urinary isolate
E. coli.
18 Interestingly, only one
P. aeruginosa isolate with
rmtA was reported in 2006.
17
Taken together, this study provides recent information regarding the 16S RMTase genes responsible for aminoglycoside-resistant isolates circulating in Korean community settings. ArmA and rmtB were the predominant 16S RMTase genes, and one RmtA-producing isolate was detected back in 2006 but not since then. Given that armA and rmtB have been found worldwide and that other 16S RMTase genes (rmtA, rmtC, rmtD, rmtF, rmtG and rmtH) are regionally spread, further studies with a larger number of clinical isolates are needed to confirm the presence of a variety of 16S RMTases among aminoglycoside-resistant bacteria in Korea.
ACKNOWLEDGMENTS
We thank Yohei Doi at University of Pittsburgh School of Medicine for critical reading and comments.
The clinical isolates for this study were provided by the Chonbuk National University Hospital, Gyeongsang National University Hospital, and Kyungpook National University Hospital, as the branches of National Culture Collection for Pathogens (NCCP).
References
1. Wachino J, Arakawa Y. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist Updat. 2012; 15(3):133–148. PMID:
22673098.
2. Galimand M, Courvalin P, Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in
Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003; 47(8):2565–2571. PMID:
12878520.
3. Galimand M, Courvalin P, Lambert T. RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob Agents Chemother. 2012; 56(7):3960–3962. PMID:
22547620.
4. Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, et al. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase,
NpmA, found in a clinically isolated
Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother. 2007; 51(12):4401–4409. PMID:
17875999.
5. Yamane K, Wachino J, Suzuki S, Shibata N, Kato H, Shibayama K, et al. 16S rRNA methylase-producing, gram-negative pathogens, Japan. Emerg Infect Dis. 2007; 13(4):642–646. PMID:
17553289.
6. Lee K, Lee M, Shin JH, Lee MH, Kang SH, Park AJ, et al. Prevalence of plasmid-mediated AmpC beta-lactamases in
Escherichia coli and
Klebsiella pneumoniae in Korea. Microb Drug Resist. 2006; 12(1):44–49. PMID:
16584308.
7. Lee H, Yong D, Yum JH, Roh KH, Lee K, Yamane K, et al. Dissemination of 16S rRNA methylase-mediated highly amikacin-resistant isolates of
Klebsiella pneumoniae and
Acinetobacter baumannii in Korea. Diagn Microbiol Infect Dis. 2006; 56(3):305–312. PMID:
16822637.
8. Park YJ, Lee S, Yu JK, Woo GJ, Lee K, Arakawa Y. Co-production of 16S rRNA methylases and extended-spectrum beta-lactamases in AmpC-producing
Enterobacter cloacae,
Citrobacter freundii and
Serratia marcescens in Korea. J Antimicrob Chemother. 2006; 58(4):907–908. PMID:
16891325.
9. Lee K, Kim MN, Kim JS, Hong HL, Kang JO, Shin JH, et al. Further increases in carbapenem-, amikacin-, and fluoroquinolone-resistant isolates of
Acinetobacter spp. and
P. aeruginosa in Korea: KONSAR study 2009. Yonsei Med J. 2011; 52(5):793–802. PMID:
21786445.
10. Citron DM, Ostovari MI, Karlsson A, Goldstein EJ. Evaluation of the E test for susceptibility testing of anaerobic bacteria. J Clin Microbiol. 1991; 29(10):2197–2203. PMID:
1939571.
11. Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007; 45(1):88–94. PMID:
17554708.
12. O'Hara JA, McGann P, Snesrud EC, Clifford RJ, Waterman PE, Lesho EP, et al. Novel 16S rRNA methyltransferase
rmtH produced by
Klebsiella pneumoniae associated with war-related trauma. Antimicrob Agents Chemother. 2013; 57(5):2413–2416. PMID:
23478957.
13. Kim MH, Sung JY, Park JW, Kwon GC, Koo SH. Coproduction of qnrB and
armA from extended-spectrum beta-lactamase-producing
Klebsiella pneumoniae
. Korean J Lab Med. 2007; 27(6):428–436. PMID:
18160833.
14. Lee HW, Kang HY, Shin KS, Kim J. Multidrug-resistant Providencia isolates carrying blaPER-1, blaVIM-2, and
armA
. J Microbiol. 2007; 45(3):272–274. PMID:
17618235.
15. Kang HY, Kim KY, Kim J, Lee JC, Lee YC, Cho DT, et al. Distribution of conjugative-plasmid-mediated 16S rRNA methylase genes among amikacin-resistant
Enterobacteriaceae isolates collected in 1995 to 1998 and 2001 to 2006 at a university hospital in South Korea and identification of conjugative plasmids mediating dissemination of 16S rRNA methylase. J Clin Microbiol. 2008; 46(2):700–706. PMID:
18094126.
16. Kang HY, Kim J, Seol SY, Lee YC, Lee JC, Cho DT. Characterization of conjugative plasmids carrying antibiotic resistance genes encoding 16S rRNA methylase, extended-spectrum beta-lactamase, and/or plasmid-mediated AmpC beta-lactamase. J Microbiol. 2009; 47(1):68–75. PMID:
19229493.
17. Jin JS, Kwon KT, Moon DC, Lee JC. Emergence of 16S rRNA methylase rmtA in colistin-only-sensitive
Pseudomonas aeruginosa in South Korea. Int J Antimicrob Agents. 2009; 33(5):490–491. PMID:
19147332.
18. Song S, Lee EY, Koh EM, Ha HS, Jeong HJ, Bae IK, et al. Antibiotic resistance mechanisms of
Escherichia coli Isolates from urinary specimens. Korean J Lab Med. 2009; 29(1):17–24. PMID:
19262074.
19. Gurung M, Moon DC, Tamang MD, Kim J, Lee YC, Seol SY, et al. Emergence of 16S rRNA methylase gene
armA and cocarriage of bla(IMP-1) in
Pseudomonas aeruginosa isolates from South Korea. Diagn Microbiol Infect Dis. 2010; 68(4):468–470. PMID:
20926221.
20. Hong SB, Shin KS, Ha J, Han K. Co-existence of blaOXA-23 and
armA in multidrug-resistant
Acinetobacter baumannii isolated from a hospital in South Korea. J Med Microbiol. 2013; 62(Pt 6):836–844. PMID:
23518656.
Table 1
16S rRNA methyltransferases in amikacin-resistant Gram-negative clinical isolates
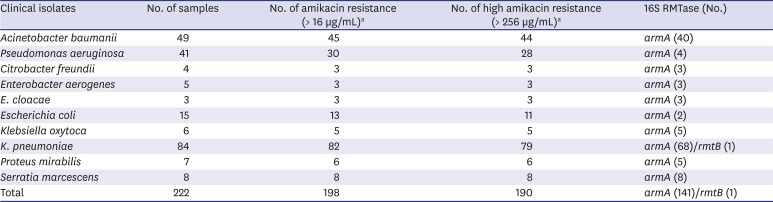
|
Clinical isolates |
No. of samples |
No. of amikacin resistance (> 16 µg/mL)a
|
No. of high amikacin resistance (> 256 µg/mL)a
|
16S RMTase (No.) |
|
Acinetobacter baumanii
|
49 |
45 |
44 |
armA (40) |
|
Pseudomonas aeruginosa
|
41 |
30 |
28 |
armA (4) |
|
Citrobacter freundii
|
4 |
3 |
3 |
armA (3) |
|
Enterobacter aerogenes
|
5 |
3 |
3 |
armA (3) |
|
E. cloacae
|
3 |
3 |
3 |
armA (3) |
|
Escherichia coli
|
15 |
13 |
11 |
armA (2) |
|
Klebsiella oxytoca
|
6 |
5 |
5 |
armA (5) |
|
K. pneumoniae
|
84 |
82 |
79 |
armA (68)/rmtB (1) |
|
Proteus mirabilis
|
7 |
6 |
6 |
armA (5) |
|
Serratia marcescens
|
8 |
8 |
8 |
armA (8) |
|
Total |
222 |
198 |
190 |
armA (141)/rmtB (1) |
Table 2
Ten published 16S rRNA methyltransferases articles related to clinical isolates from Korea
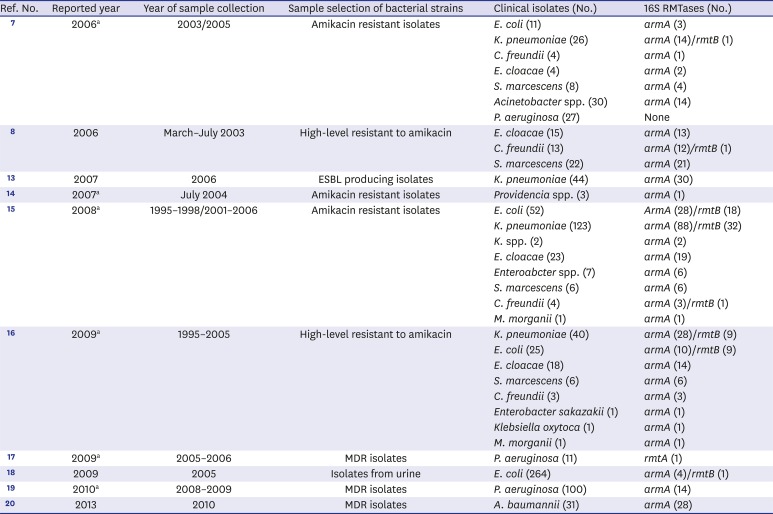
|
Ref. No. |
Reported year |
Year of sample collection |
Sample selection of bacterial strains |
Clinical isolates (No.) |
16S RMTases (No.) |
|
7
|
2006a
|
2003/2005 |
Amikacin resistant isolates |
E. coli (11) |
armA (3) |
|
K. pneumoniae (26) |
armA (14)/rmtB (1) |
|
C. freundii (4) |
armA (1) |
|
E. cloacae (4) |
armA (2) |
|
S. marcescens (8) |
armA (4) |
|
Acinetobacter spp. (30) |
armA (14) |
|
P. aeruginosa (27) |
None |
|
8
|
2006 |
March–July 2003 |
High-level resistant to amikacin |
E. cloacae (15) |
armA (13) |
|
C. freundii (13) |
armA (12)/rmtB (1) |
|
S. marcescens (22) |
armA (21) |
|
13
|
2007 |
2006 |
ESBL producing isolates |
K. pneumoniae (44) |
armA (30) |
|
14
|
2007a
|
July 2004 |
Amikacin resistant isolates |
Providencia spp. (3) |
armA (1) |
|
15
|
2008a
|
1995–1998/2001–2006 |
Amikacin resistant isolates |
E. coli (52) |
ArmA (28)/rmtB (18) |
|
K. pneumoniae (123) |
armA (88)/rmtB (32) |
|
K. spp. (2) |
armA (2) |
|
E. cloacae (23) |
armA (19) |
|
Enteroabcter spp. (7) |
armA (6) |
|
S. marcescens (6) |
armA (6) |
|
C. freundii (4) |
armA (3)/rmtB (1) |
|
M. morganii (1) |
armA (1) |
|
16
|
2009a
|
1995–2005 |
High-level resistant to amikacin |
K. pneumoniae (40) |
armA (28)/rmtB (9) |
|
E. coli (25) |
armA (10)/rmtB (9) |
|
E. cloacae (18) |
armA (14) |
|
S. marcescens (6) |
armA (6) |
|
C. freundii (3) |
armA (3) |
|
Enterobacter sakazakii (1) |
armA (1) |
|
Klebsiella oxytoca (1) |
armA (1) |
|
M. morganii (1) |
armA (1) |
|
17
|
2009a
|
2005–2006 |
MDR isolates |
P. aeruginosa (11) |
rmtA (1) |
|
18
|
2009 |
2005 |
Isolates from urine |
E. coli (264) |
armA (4)/rmtB (1) |
|
19
|
2010a
|
2008–2009 |
MDR isolates |
P. aeruginosa (100) |
armA (14) |
|
20
|
2013 |
2010 |
MDR isolates |
A. baumannii (31) |
armA (28) |






 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download