Abstract
SUV39H1 is a histone 3 lysine 9 (H3K9)-specific methyltransferase that is important for heterochromatin formation and the regulation of gene expression. Chaetocin specifically inhibits SUV39H1, resulted in H3K9 methylation reduction as well as reactivation of silenced genes in cancer cells. Histone deacetylase (HDAC) inhibitors inhibit deacetylases and accumulate high levels of acetylation lead to cell cycle arrest and apoptosis. In this study, we demonstrated that treatment with chaetocin enhanced apoptosis in human leukemia HL60, KG1, Kasumi, K562, and THP1 cells. In addition, chaetocin induced the expression of cyclin-dependent kinase inhibitor 2B (p15), E-cadherin (CDH1) and frizzled family receptor 9 (FZD9) through depletion of SUV39H1 and reduced H3K9 methylation in their promoters. Co-treatment with chaetocin and HDAC inhibitor trichostatin A (TSA) dramatically increased apoptosis and produced greater activation of genes. Furthermore, this combined treatment significantly increased loss of SUV39H1 and reduced histone H3K9 trimethylation responses accompanied by increased acetylation. Importantly, co-treatment with chaetocin and TSA produced potent antileukemic effects in leukemia cells derived from patients. These in vitro findings suggest that combination therapy with SUV39H1 and HDAC inhibitors may be of potential value in the treatment of leukemia.
Epigenetic deregulations that underlie the development of leukemia can be in one of two major categories: changes in the DNA methylation state and alterations in the histone modification pattern (1). The interaction of these two processes represents an important mechanism of epigenetic tumor suppressor gene (TSG) inactivation in the pathogenesis of human cancer (2). In the clinical trials to date, demethylation of crucial genes, such as cell cycle regulation and pro-apoptotic genes by DNA methyltransferase (DNMT) inhibitors in myeloid neoplasm is thought to be mediated through the reversal of epigenetic silencing (1, 3). However, since DNA methylation is thought to be a secondary event, and histone methylation can be a trigger in the silent state, it is possible that histone methyltransferase (HMT) inhibitors may replace DNMT inhibitors in epigenetic therapies (4, 5).
Histone 3 lysine 9 (H3K9) methylation, which was catalyzed by the histone methylase SUV39H1 and followed by the recruitment of heterochromatin protein 1 (HP), is recognized to be an inactive mark associated with transcriptional repression and heterochromatic states. In addition, H3K9 is recognized as an inactive mark associated with transcriptional repression and heterochromatic states. Interaction of SUV39H1-HP1 with histone deacetylase (HDAC) is involved in this inhibition by retinoblastoma (Rb) protein (6). Notably, SUV39H1 functions in concert with to DNA methylation via MeCP2, MBD1, and DNMT binding (7). SUV39H1 double-null mice are characterized by genomic instability and further increased risk of lymphoma in response to oncogenic Ras (8). However, its mutation is rare in epithelial cancers. Meanwhile, SUV39H1 is upregulated and associated with DNMT1 elevation in colorectal cancer (9). It was also found to be overexpressed in lung cancer cell lines, in which suppression of SUV39H1 by siRNA induced apoptosis in vitro (10). Suppression of SUV39H1 by siRNA also produced similar effects in acute myeloid leukemia (AML) cells (11, 12). In patients with an acute phase of chronic myeloid leukemia (CML) and patient with AML, strong methylation of H3K9 and all isoforms of HP1 are detected in granulocytes (13).
Epigenetic silencing of TSGs has been shown to occur in various hematopoietic neoplasms associated with cell proliferation and differentiation (2). Such as loss of p15 expression is common in AML and myeloid dysplastic syndrome (MDS) through several different mechanisms. Cancers characterized by the loss of E-cadherin (CDH1) undergo either the promoter hypermethylation or methylation independent events, which may, for example, result from the loss of a transactivating protein. Frizzled family receptor 9 (FZD9), a TSG on chromosome 7, is most frequently found in aberrantly methylated genes and its aberrant methylation combined with cytogenetic abnormalities to predict a poor clinical outcome in MDS (14). Thus, p15, CDH1, and FZD9 are TSGs that have been frequently linked to pathology in AML and MDS.
Chaetocin, a specific inhibitor of SUV39H1, potently induces cellular oxidative stress, thus selectively killing cancer cells (15-17). It has been reported to have potent anti-myeloma activity in vitro and in vivo (18). Inhibition of SUV39H1 results in reduced H3K9 methylation and enhanced expression of p15 and CDH1 in AML cell lines without promoter demethylation (11, 12). Meanwhile, the histone deacetylase inhibitor trichostatin A can reactivate gene silencing and have efficacy against leukemia in preclinical (4). Thus combined treatment with an HMT inhibitor and an HDAC inhibitor might form the optimal basis for reversing epigenetic gene inactivation and resensitizing leukemia cells to anti-tumor treatments (12, 19). Combined epigenetic therapy with the HMT inhibitor chaetocin and the HDAC inhibitor TSA has not yet been tested. In the present study, the effects of chaetocin alone and in combination with TSA, were evaluated in human leukemia cells.
Chaetocin and TSA were obtained from Sigma Aldrich (Oakville, ON, Canada). Annexin V-FITC was obtained from BD Biosciences (San Diego, CA, USA). Monoclonal anti-trimethyl histone 3 lysine 9 was obtained from Abcam (Cambridge, UK). Anti-poly (adenosine 5-diphosphate-ribose) polymerase (PARP) and anti-acetyl histone H3 lysine 9 antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). Polyclonal anti-SUV39H1 was purchased from Millipore (Temecula, CA, USA). Anti-β-Actin, normal IgG, horseradish-peroxidase conjugated secondary antibodies were acquired from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
All cell lines were obtained from American Type Culture Collection (Rockville, MD). HL60 (AML M3), KG1 (AML M6), K562 (chronic erythroleukemia in blast crisis), THP1 (AML M5), Kasumi (AML M2) carrying the t[8; 21] AML1-ETO fusion were used in this study. All cells were cultured in RPMI-1640 media supplemented with 1% penicillin/streptomycin and 10% heat inactivated fetal bovine serum (FBS) (all from Gibco-BRL, Grand Island, NY, USA) except Kasumi in media supplemented with 20% FBS. Cells were cultured in 5% CO2 at 37℃ in humidified air. Medium was changed every 2 to 3 days, and cells were passaged at a density of 0.25 × 106/mL. Logarithmically growing cell cultures were used for all experiments described below.
AML samples were obtained with informed consent in accordance with the Declaration of Helsinki as part of a clinical protocol approved by the Institutional Review Board of the Chonnam National University Hwasun Hospital. Fresh bone marrow (BM) aspirate samples at diagnosis were collected and separated for mononuclear cells, as previous described (19). 1 × 106 cells were incubated in six-well plates in the presence or absent of 100 nM of chaetocin with or without 1 µM of TSA for 18 hr at 37℃ in 5% CO2 humidified air. The drug concentrations were selected based on our results of cell lines.
For cytometric evaluation of apoptosis in AML cell lines, untreated and drug treated cells were washed with cold PBS and resuspended in 1 × Binding buffer. Cells were stained by incubated with 5 µL of annexin V (Pharmingen, San Diego, CA, USA) and 10 µL of propidium iodide (PI) (Invitrogen) for 15 min in the dark at room temperature, and the percentage of apoptotic cells were determined by flow cytometry within 1 hr.
Western blotting analysis of total protein lysates was performed as previously described (19). Briefly, cell pellets were washed twice with 1 × PBS, resuspended in RIPA buffer supplemented with a protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 µg/mL leupeptin, 1 µg/mL pepstatin-A, 1 µg/mL aprotinin, 1 mM sodium orthovanadate, and 1 mM sodium fluoride), and incubated on ice. Cell lysates were centrifuged at 12,000 rpm for 15 min to remove the nuclear and cellular debris. Protein concentration was determined with the BCA assay (Pierce Chemical, West Pico, Rockville, IL, USA).
One hundred micrograms of total cell lysate were separated by SDS-PAGE and blotted onto a membrane. The membranes were blocked in blocking solution, incubated with primary antibodies (e.g. PARP) over-night, and secondary antibodies for 1-2 hr. After that, the blots were developed by using a two-component ECL detection reagent (GE Healthcare, Buckinghamshire, UK) and exposed to scientific imaging film. Immunoblot analyses were performed at least twice, and representative blots were subjected to densitometric analysis. Densitometry was performed using Multi Gauge v3.2 software (Fuji film Corporation, Tokyo, Japan).
Total RNA was harvested using TRIzol (Invitrogen) according to the manufacturer's instructions and reverse transcribed using a SuperScript First-Strand Synthesis kit (Invitrogen). All samples were plated in triplicate, and then loaded onto a 72-well Rotor-Gene RG-3000 (Corbett Research, Sydney, Australia) with a 10 µL final reaction mixture containing 250 nM of each primer, 1 × SYBR Green (Takara, Tokyo, Japan), and cDNA. The primer sequences are listed in Table 1. The reaction mixture was preheated to 95℃ for 10 sec, followed by 45 cycles of 95℃ for 10 sec, 60℃ for 20 sec, and 72℃ for 20 sec. All reactions included negative controls where reverse transcriptase was omitted.
Chromatin immunoprecipitation (ChIP) assay were performed, according to the manufacturer's protocol (Upstate Biotechnology, Temecula, CA, USA). The primary antibody (anti trimethyl H3K9) and rabbit IgG (negative control) were used. For quantitative assessment of p15, CDH1, and FZD9 in the chromatin immunoprecipitates, real time PCR using SYBR Green (Takara, Tokyo, Japan) was performed. Amplified products were normalized to the non-specific glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter enrichment. Relative enrichment in the chromatin immunoprecipitates was normalized against p15, CDH1, and FZD9 in the input samples. Primer sequences are listed in Table 1.
Histones were isolated by a modification of a previously described method (20). After the designated treatments, cells were harvested and incubated on ice cold histone isolation buffer (PBS containing 0.5% Triton X-100 (v/v), 2 mM phenylmethylsulfonyl fluoride, and 0.02% NaN3) on ice for 10 min with gentle stirring. Nuclei were pelleted by centrifuge at 6,500 × g for 10 min at 4℃ and washed briefly with the histone lysis buffer. The nuclei were resuspended in 0.2 N HCl and the tubes were incubated overnight at 4℃. The acid-treated nuclei were centrifuged for 10 min at 6,500 × g. The supernatants were removed to a clean microcentrifuge tube, and 1 mL acetone was added. Histones were precipitated from the acid extracts overnight at -20℃. The extracted histones were centrifuged briefly at 10,000 rpm, air dried, and resuspended in 50 µL water. Protein concentrations were quantified as described above. For Western blot, 1 to 3 µg of purified histones were used per condition.
All statistical analyses were performed with the program SPSS 13.0 for Windows. Significant differences between values obtained in a population of leukemic cells treated with different experimental conditions were determined using the Mann-Whitney U-test. P values < 0.05 were assigned significance.
In the earlier studies, chaetocin had a cytotoxic effect on Drosophila cell (15), and on myeloma (18). Chaetocin and SUV39H1 shRNA substantially increased cell cycle arrest in human leukemia AML-193, KG1, and U937 cells (11, 12), as well as microglial cells (21). In this study, we first assessed the biologic effect of chaetocin on different representative cell lines-HL60, KG1, Kasumi, K562, and THP1-on apoptosis by Annexin V staining. Treatment of these cells with increasing doses of chaetocin (0-500 nM) for 24 hr induced greater apoptosis (Fig. 1A). Next, these results were confirmed by Western blotting after treatment with chaetocin for 24 hr. Exposure to chaetocin dose dependently induced caspase-dependent cleavage of PARP to a greater extent in myeloid cells (Fig. 1B). Moreover, chaetocin induced apoptosis in a time dependent manner (Fig. 1C).
Re-expression of epigenetically silenced TSGs as a result of SUV39H1 inhibition has been reported (11). We ourselves previously described increased p15 and CDH1 mRNA expression in KG1 and Kasumi cells. We next determined the effects of various concentrations of chaetocin treatment on expression of the p15, CDH1, and FZD9 genes in the myeloid cell lines. The results showed that treatment with 100-200 nM chaetocin resulted in strong re-expression of epigenetically silenced/weakly expressed p15, CDH1 and FZD9 genes in HL60, KG1, and Kasumi cells, as well as re-expression of CDH1 and FZD9 in K562 and THP1 cells (Fig. 2) (P<0.05). They also revealed p15 deletions in K562 and THP1 cells, as indicated by the lack of a p15 signal in those cell lines, according to the characteristic of these cell lines.
In Drosophila SL-2, chaetocin has been demonstrated to deplete the activity of SUV39H1 (15). Consistent with this report, treatment of HL60, KG1, Kasumi, K562 and THP1 myeloid cells with chaetocin dose-dependently reduced SUV39H1 protein levels (Fig. 3A), which can lead to the inhibition of H3K9 methylation. Recently, chaetocin was shown to reduce SUV39H1 and H3K9 trimethylation in the promoter regions of the p21 (21), p15, and CDH1 (11) genes. In our study, ChIP assays were performed using anti-trimethyl-H3K9 to analyze the effect of chaetocin on the p15, CDH1 and FZD9 promoters in these cell lines. The result showed that the levels of trimethylation of H3K9 in the p15, CDH1 and FZD9 promoter regions decreased relative to the untreated control cells in HL60, KG1 and Kasumi cells (Fig. 3B). Also, this association with the CDH1 and FZD9 promoters was down-in regulated K562, and THP1 cells compared to untreated control cells (Fig. 3B).
The IC50 values of the HDAC inhibitor TSA for apoptosis in different cell lines were determined by flow cytometry (data not shown). The concentration 1 µM of TSA caused cell death in these cells. To determine whether TSA enhances the effects of chaetocin in leukemia cell lines, the effects of the combination of the two compounds on apoptosis were evaluated. As shown in Fig. 4A, apoptosis was higher in myeloid cell lines treated with both chaetocin and TSA than in cells treated with chaetocin alone. These responses were accompanied by synergistically increased PARP cleavage as verified by Western blotting (Fig. 4B), indicating that the antileukemic activity of the combined treatment was higher than that of the individual compounds. Additionally the combination of compounds caused more apoptosis in HL60, Kasumi, and K562 cells than in KG1 or THP1 cells (Fig. 4A).
In HL60, K562, and particularly in Kasumi cells, the combination of chaetocin and TSA produced a markedly stronger re-expression of these genes than that achieved through treatment with chaetocin alone (Fig. 4C). In addition, KG1 cells showed a significant increase in p15 expression. In contrast, combined treatment somehow did not increase activation of CDH1 and FZD9 genes in KG1 or THP1 cells, the results exposed to higher than untreated controls (Fig. 2 and 4C). Taken together, these findings can support greater activation of silencing genes in leukemia cells treated with a combination of chaetocin and TSA than in cells treated with chaetocin alone.
A link between the growth inhibitory and apoptotic antileukemic activities that are associated with epigenetic inhibition and the reactivation of genes has been identified (19). As shown in Fig. 5A, although TSA reduced histone methyltransferase levels in some myeloid cells, co-treatment with chaetocin and TSA produced a stronger reduction in SUV39H1 protein levels than treatment with either agent alone. Treatment of HL60, KG1, Kasumi, K562, and THP1 cells with a combination of chaetocin and TSA more strongly reduced histone H3K9 trimethylation, and more strongly induced increased histone H3K9 acetylation, than treatment with chaetocin alone (Fig. 5B).
The effects of chaetocin and TSA were assessed in leukemia cells isolated from fresh BM from seven patients with AML. The characteristics of the patients are summarized in Table 2. As shown in Fig. 6A, chaetocin killed primary AML cells through induction of PARP cleavage and reduced SUV39H1 protein levels in a dose-dependent manner. The expression of SUV39H1 under the treatment of chaetocin at a concentration of 100 nM seem to be higher than no treatment, however, the densitometric analysis by Multi Gauge v3.2 software reveals that the expression of SUV39H1 was decreased in a dose-dependent manner. In addition, combined treatment with chaetocin and TSA induced greater PARP cleavage and loss of SUV39H1 protein in AML cells than single agent alone. As shown in Fig. 6B, as measured by annexin V staining, indicating the stronger antileukemic activity of this combined treatment compared to that of the individual agents; although levels of apoptosis in normal leukocytes were no increase in the combination (data not shown). Note that all AML samples were sensitive to combined treatment, despite the heterogeneity of their biologic features: patients 1, 4, 6, and 7 were newly diagnosed with FAB M2; patient 2 had a mixed leukemia (T/myeloid); patient 3 had secondary AML arising from a cancer treated with chemoradiotherapy for 9 yr; and patient 5 had M4. Cells from the patients with M2 (P1, 4, 6, and 7), blast >85% (P2, 4, and 7) or AML1-ETO fusion (P1) were more effectively killed by the combined treatment. Interestingly, the effect of combine treatment was observed not only in primary AML but also effective in secondary AML (P3). Concerning patient mutation status, only patient 6 carried three out of four gene mutations screened (FLT3-ITD, C/EBPα and NPM1). In addition, combined analysis of the activity of chaetocin and TSA in the total cells from the seven patients showed the combined treatment to be significantly superior to the individual agents (Fig. 6C).
Chaetocin was significantly more potent in myeloid cells, as indicated by its dose- and time-dependent enhancement of apoptosis in human leukemia cells. Chaetocin induces apoptosis in leukemia cell lines in vitro and primary AML cells ex vivo has been reported (12). Identification of an optimal combination therapy, in particular epigenetic th erapy is ongoing. To our knowledge, our study is the first to describe the combined effects of the HMT inhibitor chaetocin and the HDAC inhibitor TSA on apoptosis. Moreover, chaetocin was also effective in AML cells from patients, particularly when co-administered with TSA.
As reported previously, chaetocin upregulates the transcription of death-receptor-related genes, leading to death receptor-dependent apoptosis (12, 17). Pro-apoptotic activity of chaetocin may be mediated, at least in part, by inhibition of SUV39H1 HMT activity at the TSG promoter (11, 21). Overexpression of SUV39H1 mRNA was found in AML cell lines (data not shown). Treatment of leukemia cell lines with the SUV39H1 inhibitor chaetocin reduced SUV39H1 levels and lowered H3K9 methylation in the p15, CDH1, and FZD9 promoters, thereby reactivating their expression. Furthermore, we demonstrated that chaetocin-mediated depletion of SUV39H1 was associated with increased apoptosis in AML cells from patients. The development of aberrant TSG silencing may stem from increased SUV39H1 binding and H3K9 trimethylation in their promoters as a result of interactions with DNA-binding proteins (5). A possible candidate for targeting SUV39H1 to the p15, Evi1 interacts with SUV39H1 and represses TGF-β-induced activation of the p15 promoter (22). CDH1 promoter contains AML1-binding sites, and thus AML1 may recruit SUV39H1 to repress CDH1 expression (23). Through methyl-H3K9 binding, MPP8 targets CDH1 gene promoter and modulates CDH1 gene expression (24). FZD9 is activated by Wnt-2 and functions in Wnt signaling (25). Although the mechanism by which recruitment of SUV39H1 inhibits FZD9 expression remains unknown, our findings demonstrate that chaetocin reduced SU39H1 protein, thereby reducing H3K9 methylation in the FZD9 promoter and resulting in FZD9 re-expression in leukemia cell lines.
Given the multiple effects of HDAC inhibitors on malignant cells, their true therapeutic potential most likely lies in combination with other anticancer drugs (26). TSA is an antileukemic agent that has been reported to be a potent inducer or enhancer of differentiation in AML (27). We therefore focused on the anticancer effect of the HMT inhibitor chaetocin in combination with the HDAC inhibitor TSA. Interestingly, high levels of apoptosis (at least 50% apoptotic cells) was detected in TSA-treated HL60, Kasumi and K562 cells, but not similarly treated KG1 or THP1 cells. So, while TSA showed potential as a single agent, combined treatment with chaetocin and TSA produced significantly stronger effects in HL60, Kasumi, and K562 cells compared to KG1 and THP1 cells (Fig. 4A). Notably, the effect of these compounds on apoptosis was outstanding in Kasumi cells. About 70% apoptosis was observed in cells treated with individual agent, while apoptosis was almost maximal in cells treated with both agents. This may have been due to the fact that Kasumi cells carry an AML1-ETO fusion that predisposes myeloid precursors to transformation, and that turnover of the myeloid oncoprotein was induced by TSA (28). Although TSA dramatically degrades AML1-ETO fusion prior to inducing apoptosis in Kasumi cells (28), additional mechanisms underlying the induction of apoptosis by TSA and chaetocin in t(8;21) AML cell lines should be further investigated. Nevertheless, these finding also suggest that chaetocin and/or TSA may be a potent targeted therapies for leukemia, especially t(8;21) AML, as they target multiple pathways, including those inducing apoptosis.
Re-expression of epigenetically inactivated genes can result in the suppression of tumor growth or a sensitization to anticancer therapy. Compared to either agent alone, co-treatment with chaetocin and TSA produced greater responses. Therefore, chaetocin and TSA cooperated in inducing TSG expression, which may potentially have contributed to the superior antileukemic activity of the combined treatment in HL60, Kasumi and K562 cells compared to KG1 and THP1 cells. Simultaneously, the greater decline in trimethylated H3K9 levels and increase in hyperacetylated H3K9 levels were found to relieve gene repression. Notably, the induction of apoptosis by the combination of chaetocin and TSA was correlated with the increased expression of these genes, in HL60, Kasumi and K562 compared to KG1 and THP1 cells (Fig. 4C). The combination of TSA and chaetocin may induce transcriptional activation and activity of apoptotic proteins, thereby sensitizing cells to apoptosis (26). In addition, the AML1-ETO fusion acts to repress the transcription of key regulatory genes such as C/EBPα, which regulates the cell cycle control and myeloid differentiation (29). Inhibitors target this oncoprotein, resulting in greater derepression and cancer cell death. Kasumi cells co-treated with TSA and chaetocin exhibited greater expression of p15, CDH1, and FZD9 than similarly treated HL60 and K562 cells. The latter is known to trigger cell death though multiple mechanisms in transformed versus normal hematopoietic progenitor cells via ROS production selectively in cancer cells (12, 16-18). The p15 gene regulates Rb function by modulating cyclin D-CDK4/6 complexes, thereby inhibiting cell growth (30). The expression of CDH1 were decreased during tumor progression and metastasis in a variety of cancers (31, 32). The loss of FZD9 in acute lymphoblastic leukemia was associated with increased in cell proliferation and cell cycle entry (33). Chaetocin may modulate several of these mechanisms to increase apoptosis in response to TSA treatment. Reduced p53-triggered transactivation of p21 allows cells to reenter the cell cycle. TSA activates p53, thereby inducing apoptosis in colon cancer cells (34). TSA upregulates p73 and induces Bax-dependent apoptosis in ovarian cancer cells (35).
Consistent with the published reports (12, 18, 36, 37), chaetocin and TSA has little effect in non-cancer cells, suggesting cancer-cell-specific cytotoxicity. In conclusion, the present study demonstrated that a combination of the HMT inhibitor chaetocin and the HDAC inhibitor TSA may provide effective treatment and enhance TSG reactivation in epigenetic therapies in leukemia cells. The reversal of SUV39H1-mediated gene repression by chaetocin in leukemia still needs to be clarified. In addition, more selective HDAC inhibitors should be tested. Recently, similar combined epigenetic therapy with an HMT EZH2 inhibitor and an HDAC inhibitor was reported to be effective in AML cells (19).
Taken together, the findings presented in this study provide the rationale to design and implement further studies to investigate the in vivo antileukemic effects of combined chaetocin and TSA treatment.
Figures and Tables
Fig. 1
Chaetocin induces apoptosis in the leukemia cell lines. (A) After treated 24 hr, apoptotic cells were determined by flow cytometry. (B) Confirmatory Western blotting. (C) After treated 48 and 72 hr. Beta actin is protein loading control. C-48: 48 hr-untreated control, C-72: 72 hr-untreated control. *P < 0.05.

Fig. 2
Real-time PCR analysis of p15, CDH1 and FZD9 mRNA expression in HL60, KG1, Kasumi, K562 and THP1 cells. Gene expression was normalized to β2-microglobin. The expression of p15, CDH1 and FZD9 in chaetocin-treated cells is shown relative to that in untreated cells. *P < 0.05; †P < 0.01.

Fig. 3
Chaetocin reduces SUV39H1 protein levels and H3K9 methylation in p15, CDH1 and FZD9 promoters. (A) Western blotting was showed with beta actin as a loading control. (B) The effect of chaetocin (100 nM) on H3K9 trimethylation in the promoters in HL60, KG1, Kasumi, K562 and THP1 cells was analyzed by ChIP assays using anti-trimethyl H3K9 (H3K9trime). Histograms show antibody/input ratios for PCR products, quantified by real-time PCR. *P < 0.05.

Fig. 4
Chaetocin and TSA co-treatmentlly induce apoptosis and gene expression after 24 hr treatment in myeloid leukemia cell lines. (A) Apoptosis was determined by flow cytometry. (B) Confirmatory Western blotting with beta actin as a loading control. (C) Real-time PCR of p15, CDH1 and FZD9 in chaetocin and TSA-treated cells is shown relative to that in chaetocin-treated cells. *P < 0.05; †P < 0.01; Ch100: Chaetocin concentration at 100 nM.
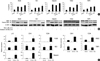
Fig. 5
Co-treatment with chaetocin and TSA alter the expression of methyltransferase related molecules. Myeloid cell lines were harvested after 24 hr incubation with indicated drug. Western blotting analyses were performed and representative blots were subjected to densitometric analysis. Beta actin was used as a loading control for cell lysates. Ponceau-stained histones were used as a loading control for acid-extracted histones. (A) Combined treatment with chaetocin and TSA increases SUV39H1 protein depletion. (B) The combined treatment reduces histone H3K9 trimethylation, and increases histone H3K9 acetylation. H3K9 trime: histone H3K9 trimethylation; H3K9 ac: histone H3K9 acetylation.

Fig. 6
Co-treatment with chaetocin and TSA produces stronger antileukemic effects in cells from patients with AML. (A) Western blotting analyses were performed and representative blots were subjected to densitometric analysis. (B) Apoptosis of cells from bone marrow seven patients with AML. (C) Box plots showing the percentage of apoptotic cells in the AML cell populations of the seven analyzed patients after treatment with the indicated doses. *P < 0.01. Pa: patient; Ch100 nM: Chaetocin concentration at 100 nM.

Notes
This study was supported by a grant of the National Project for Personalized Genomic Medicine, Ministry for Health & Welfare, Republic of Korea (A111218-GM06) and a grant of the Environmental Health Center for Childhood Leukemia and Cancer, Chonnam National University Hwasun Hospital, Republic of Korean (HCRE/0035-6).
References
1. Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010. 10:23–36.
2. Krug U, Ganser A, Koeffler HP. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene. 2002. 21:3475–3495.
3. Tran HT, Kim HN, Lee IK, Kim YK, Ahn JS, Yang DH, Lee JJ, Kim HJ. DNA methylation changes following 5-azacitidine treatment in patients with myelodysplastic syndrome. J Korean Med Sci. 2011. 26:207–213.
4. Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004. 4:143–153.
5. Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002. 14:286–298.
6. Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002. 30:475–481.
7. Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem. 2003. 278:24132–24138.
8. Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, DÖrken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005. 436:660–665.
9. Kang MY, Lee BB, Kim YH, Chang DK, Park SK, Chun HK, Song SY, Park J, Kim DH. Association of the SUV39H1 histone methyltransferase with the DNA methyltransferase 1 at mRNA expression level in primary colorectal cancer. Int J Cancer. 2007. 121:2192–2197.
10. Watanabe H, Soejima K, Yasuda H, Kawada I, Nakachi I, Yoda S, Naoki K, Ishizaka A. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer Cell Int. 2008. 8:15.
11. Lakshmikuttyamma A, Scott SA, DeCoteau JF, Geyer CR. Reexpression of epigenetically silenced AML tumor suppressor genes by SUV39H1 inhibition. Oncogene. 2010. 29:576–588.
12. Chaib H, Nebbioso A, Prebet T, Castellano R, Garbit S, Restouin A, Vey N, Altucci L, Collette Y. Anti-leukemia activity of chaetocin via death receptor-dependent apoptosis and dual modulation of the histone methyl-transferase SUV39H1. Leukemia. 2012. 26:662–674.
13. Lukásová E, Koristek Z, Falk M, Kozubek S, Grigoryev S, Kozubek M, Ondrej V, Kroupová I. Methylation of histones in myeloid leukemias as a potential marker of granulocyte abnormalities. J Leukoc Biol. 2005. 77:100–111.
14. Jiang Y, Dunbar A, Gondek LP, Mohan S, Rataul M, O'Keefe C, Sekeres M, Saunthararajah Y, Maciejewski JP. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009. 113:1315–1325.
15. Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005. 1:143–145.
16. Tibodeau JD, Benson LM, Isham CR, Owen WG, Bible KC. The anticancer agent chaetocin is a competitive substrate and inhibitor of thioredoxin reductase. Antioxid Redox Signal. 2009. 11:1097–1106.
17. Isham CR, Tibodeau JD, Bossou AR, Merchan JR, Bible KC. The anticancer effects of chaetocin are independent of programmed cell death and hypoxia, and are associated with inhibition of endothelial cell proliferation. Br J Cancer. 2012. 106:314–323.
18. Isham CR, Tibodeau JD, Jin W, Xu R, Timm MM, Bible KC. Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood. 2007. 109:2579–2588.
19. Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009. 114:2733–2743.
20. Fiskus W, Pranpat M, Balasis M, Herger B, Rao R, Chinnaiyan A, Atadja P, Bhalla K. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol Cancer Ther. 2006. 5:3096–3104.
21. Cherrier T, Suzanne S, Redel L, Calao M, Marban C, Samah B, Mukerjee R, Schwartz C, Gras G, Sawaya BE, et al. P21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene. 2009. 28:3380–3389.
22. Spensberger D, Delwel R. A novel interaction between the proto-oncogene Evi1 and histone methyltransferases, SUV39H1 and G9a. FEBS Lett. 2008. 582:2761–2767.
23. Chakraborty S, Sinha KK, Senyuk V, Nucifora G. SUV39H1 interacts with AML1 and abrogates AML1 transactivity: AML1 is methylated in vivo. Oncogene. 2003. 22:5229–5237.
24. Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010. 29:3673–3687.
25. Karasawa T, Yokokura H, Kitajewski J, Lombroso PJ. Frizzled-9 is activated by Wnt-2 and functions in Wnt/beta -catenin signaling. J Biol Chem. 2002. 277:37479–37486.
26. Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009. 15:3970–3977.
27. Kosugi H, Towatari M, Hatano S, Kitamura K, Kiyoi H, Kinoshita T, Tanimoto M, Murate T, Kawashima K, Saito H, et al. Histone deacetylase inhibitors are the potent inducer/enhancer of differentiation in acute myeloid leukemia: a new approach to anti-leukemia therapy. Leukemia. 1999. 13:1316–1324.
28. Yang G, Thompson MA, Brandt SJ, Hiebert SW. Histone deacetylase inhibitors induce the degradation of the t(8;21) fusion oncoprotein. Oncogene. 2007. 26:91–101.
29. Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, Hiddemann W, Zhang DE, Tenen DG. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001. 7:444–451.
30. Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996. 85:27–37.
31. Karayiannakis AJ, Syrigos KN, Chatzigianni E, Papanikolaou S, Alexiou D, Kalahanis N, Rosenberg T, Bastounis E. Aberrant E-cadherin expression associated with loss of differentiation and advanced stage in human pancreatic cancer. Anticancer Res. 1998. 18:4177–4180.
32. Sulzer MA, Leers MP, van Noord JA, Bollen EC, Theunissen PH. Reduced E-cadherin expression is associated with increased lymph node metastasis and unfavorable prognosis in non-small cell lung cancer. Am J Respir Crit Care Med. 1998. 157:1319–1323.
33. Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br J Haematol. 2007. 138:338–348.
34. Habold C, Poehlmann A, Bajbouj K, Hartig R, Korkmaz KS, Roessner A, Schneider-Stock R. Trichostatin A causes p53 to switch oxidative-damaged colorectal cancer cells from cell cycle arrest into apoptosis. J Cell Mol Med. 2008. 12:607–621.
35. Muscolini M, Cianfrocca R, Sajeva A, Mozzetti S, Ferrandina G, Costanzo A, Tuosto L. Trichostatin A up-regulates p73 and induces Bax-dependent apoptosis in cisplatin-resistant ovarian cancer cells. Mol Cancer Ther. 2008. 7:1410–1419.
36. Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006. 6:38–51.
37. Zhang Y, Yu G, Wang D, Hu Y, Lei W. ERK1/2 activation plays important roles in the opposite effects of Trichostatin A in non-cancer and cancer cells. Toxicon. 2011. 57:932–937.




 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download