Abstract
Background
Extraction of cell-free DNA (cfDNA) is a key step for determining the quality of cfDNA-related molecular diagnostics. We evaluated the effect of sample containers and sample storage conditions on cfDNA extraction.
Methods
The cfDNA extraction using the MagMAX Cell-Free DNA Isolation Kit from five healthy controls and five lung cancer patients was evaluated according to the type of sample container and storage conditions: K2-EDTA container, <1, 6, 24, and 48 hr storage at 4˚C after immediate plasma separation; and Cell-Free DNA BCT container, <1, 3, 7, and 14 days stored at room temperature. Mutation analysis of EGFR exons 18–21 was performed. To assess the effect of a delay in centrifugation, EDTA whole blood samples from five healthy individuals were stored at 4˚C for 6, 12, and 24 hr before plasma separation.
Results
There was no significant difference in the amount and nucleic acid size of cfDNA in both controls and patients with cancer when EDTA plasma was stored at 4˚C up to 48 hr. The amount and size of cfDNA in the BCT container were not different up to 7 days; however, the 14-day sample showed an increase in cfDNA concentration due to genomic DNA contamination. EGFR mutations were detected on EDTA containers up to 48 hr and with BCT containers up to 14 days. When EDTA whole blood was stored at 4˚C and plasma separation was delayed, the cfDNA concentration increased from 24 hr.
References
1. Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): Clinical signifcance and utility in cancer shaped by emerging technologies. Mol Cancer Res. 2016; 14:898–908.
2. Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Cal-das C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017; 17:223–38.

3. Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008; 359:366–77.
4. Kwapisz D. The frst liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 2017; 5:46.
5. Diaz LA Jr and Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014; 32:579–86.
6. Ilie M and Hofman P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res. 2016; 5:420–3.
7. Krishnamurthy N, Spencer E, Torkamani A, Nicholson L. Liquid biopsies for cancer: Coming to a patient near you. J Clin Med. 2017; 6:3.

8. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012; 366:883–92.

9. Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013; 501:355–64.

10. Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016; 35:347–76.

11. Yao W, Mei C, Nan X, Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene. 2016; 590:142–8.

12. Bronkhorst AJ, Aucamp J, Pretorius PJ. Cell-free DNA: Preanalytical variables. Clin Chim Acta. 2015; 450:243–53.

13. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015; 61:112–23.

14. Chen W, Cai F, Zhang B, Zhong XY. Strategies of reducing input sample volume for extracting circulating cell-free nuclear DNA and mitochondrial DNA in plasma. Clin Chem Lab Med. 2011; 50:261–5.

15. Devonshire AS, Whale AS, Gutteridge A, Jones G, Cowen S, Foy CA, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction effciency, fragment size bias and quantifcation. Anal Bioanal Chem. 2014; 406:6499–512.
16. van Dessel LF, Beije N, Helmijr JC, Vitale SR, Kraan J, Look MP, et al. Application of circulating tumor DNA in prospective clinical oncology trials – standardization of preanalytical conditions. Mol Oncol. 2017; 11:295–304.

17. Medina Diaz I, Nocon A, Mehnert DH, Fredebohm J, Diehl F, Holtrup F. Performance of Streck cfDNA blood collection tubes for liquid biopsy testing. PLoS One. 2016; 11:e0166354.

18. Sherwood JL, Corcoran C, Brown H, Sharpe AD, Musilova M, Kohl-mann A. Optimised preanalytical methods improve KRAS mutation detection in circulating tumour DNA (ctDNA) from patients with nonsmall cell lung cancer (NSCLC). PLoS One. 2016; 11:e0150197.

19. Parpart-Li S, Bartlett B, Popoli M, Adleff V, Tucker L, Steinberg R, et al. The effect of preservative and temperature on the analysis of circulating tumor DNA. Clin Cancer Res. 2017; 23:2471–7.

20. van Ginkel JH, van den Broek DA, van Kuik J, Linders D, de Weger R, Willems SM, et al. Preanalytical blood sample workup for cell-free DNA analysis using Droplet Digital PCR for future molecular cancer diagnostics. Cancer Med. 2017; 6:2297–307.

21. Warton K, Yuwono NL, Cowley MJ, McCabe MJ, So A, Ford CE. Evaluation of Streck BCT and PAXgene stabilised blood collection tubes for cell-free circulating DNA studies in plasma. Mol Diagn Ther. 2017; 21:563–70.

22. Streck. Cell-Free DNA BCT: INSTRUCTIONS FOR USE. Omaha N, USA;. 2014. https://www.streck.com/collection/cell-free-dna-bct/. (Assessed on Jan 2018).
23. Alix-Panabieres C and Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016; 6:479–91.

24. Zhang R, Peng R, Li Z, Gao P, Jia S, Yang X, et al. Synthetic circulating cell-free DNA as quality control materials for somatic mutation detection in liquid biopsy for cancer. Clin Chem. 2017; 63:1465–75.

25. Xue X, Teare MD, Holen I, Zhu YM, Woll PJ. Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta. 2009; 404:100–4.

26. Leung F, Kulasingam V, Diamandis EP, Hoon DS, Kinzler K, Pantel K, et al. Circulating tumor DNA as a cancer biomarker: fact or fction? Clin Chem. 2016; 62:1054–60.
Fig. 1.
Schematic flow of the study design. (A) Venous whole blood (WB) was drawn from 10 subjects (healthy controls, N=5; lung cancer patients, N=5). The cell-free DNA (cfDNA) was extracted under different specimen storage conditions in K2-EDTA at 4°C and Cell-Free DNA BCT containers at room temperature (RT). (B) Venous blood was drawn from five healthy subjects in K2-EDTA containers, and the cfDNA was extracted under different centrifugation conditions.
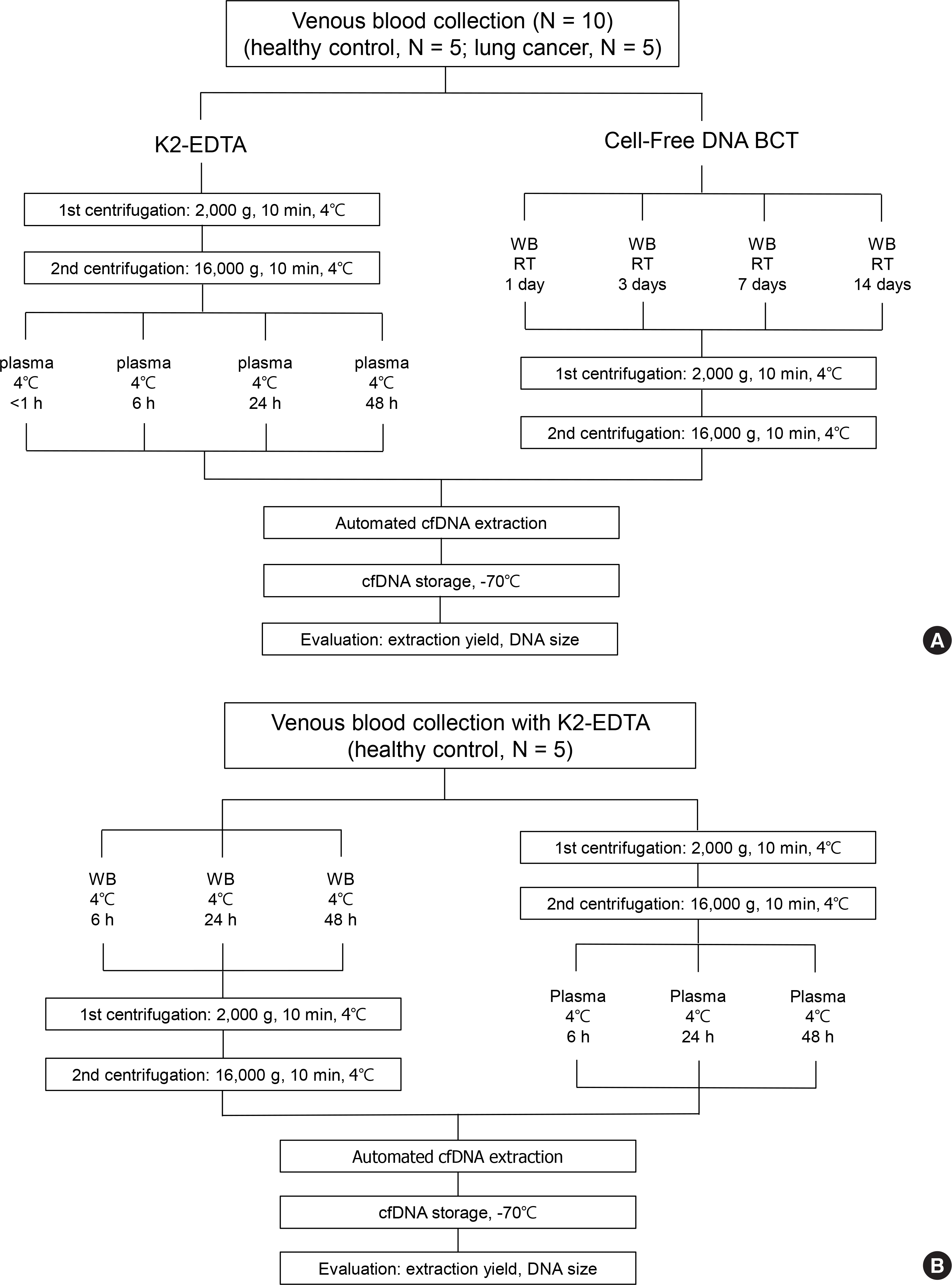
Fig. 2.
Comparison of cell-free DNA (cfDNA) concentrations according to specimen container in healthy controls and patients with lung cancer. The cfDNA concentration of patients with lung cancer was significantly higher than that of healthy controls. In the healthy control group, the mean concentration of cfDNA in the K2-EDTA container was significantly higher than that of the Cell-Free DNA BCT containers.
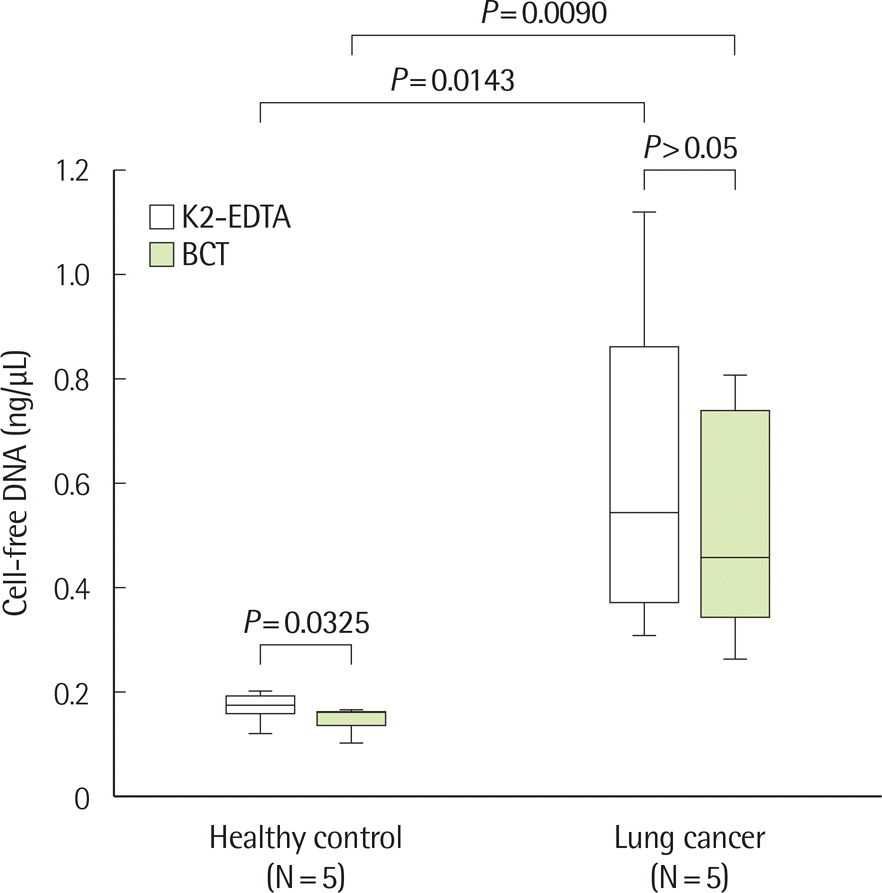
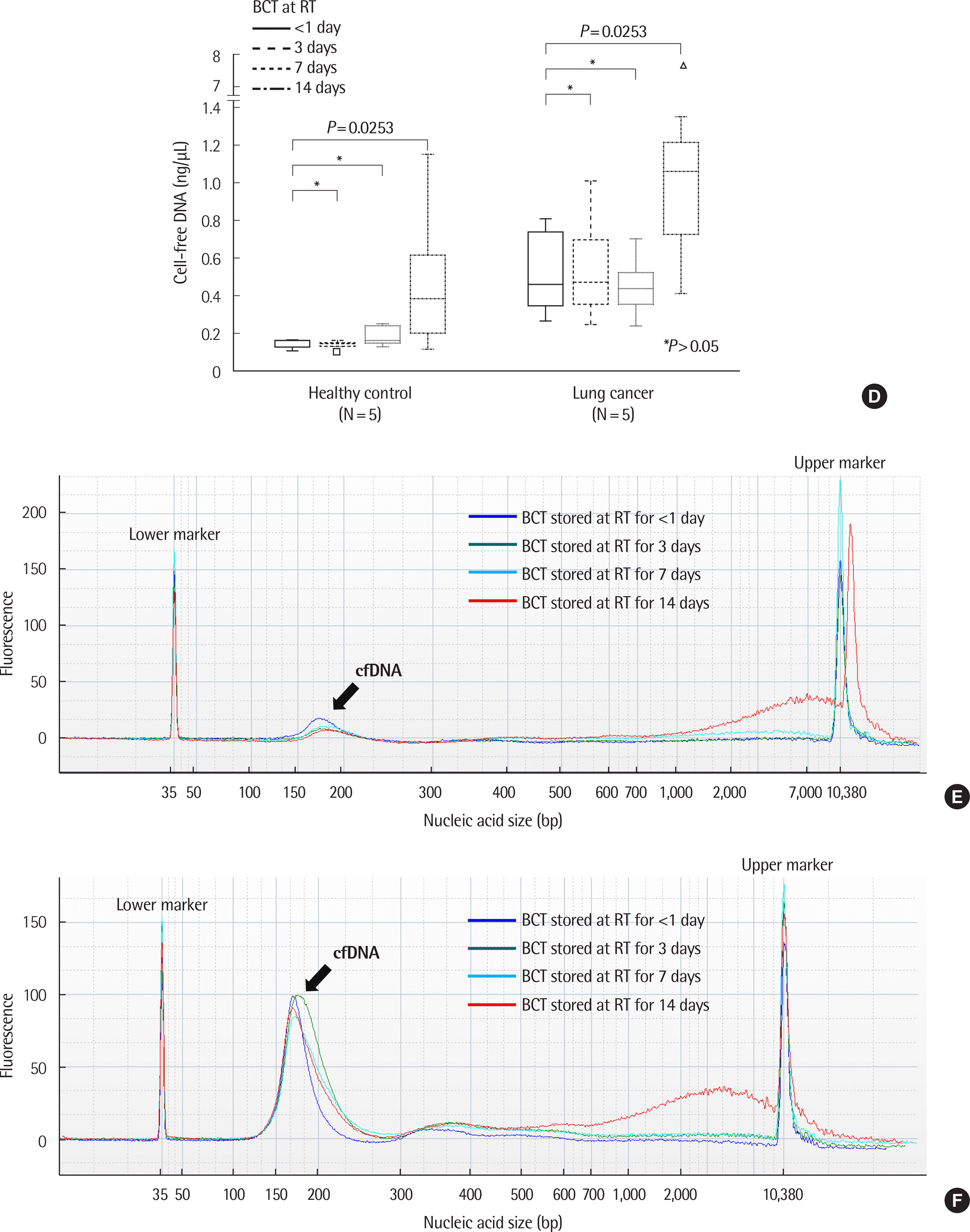
Fig. 3.
Comparison of cfDNA concentration and size according to storage conditions in healthy controls and patients with lung cancer. (A) Changes in cfDNA concentration over time in EDTA plasma. There were no significant differences in cfDNA concentration up to 48 hr in EDTA plasma stored at 4˚C. ∗P >0.05 (one way repeated measures ANOVA or Friedman test, N=15). (B) Example of size distribution of nucleic acids in EDTA plasma in a healthy control (case number: HC04). No significant differences were observed according to time. (C) Example of size distribution of nucleic acids on EDTA plasma in a patient with lung cancer (case number: LC09). No significant differences were observed according to time. (D) Changes of cfDNA concentrations over time in BCT containers. There were no statistically significant differences in cfDNA concentration up to 14 days in BCT stored at room temperature (RT). On day 14, an increase in cfDNA concentration was observed. ∗P >0.05 (one way repeated measures ANOVA or Friedman test, N=15). (E) Example of size distribution of nucleic acids in BCT in a healthy control (case number: HC04). (F) Example of size distribution of nucleic acids in BCT in a lung cancer patient (case number: LC09).
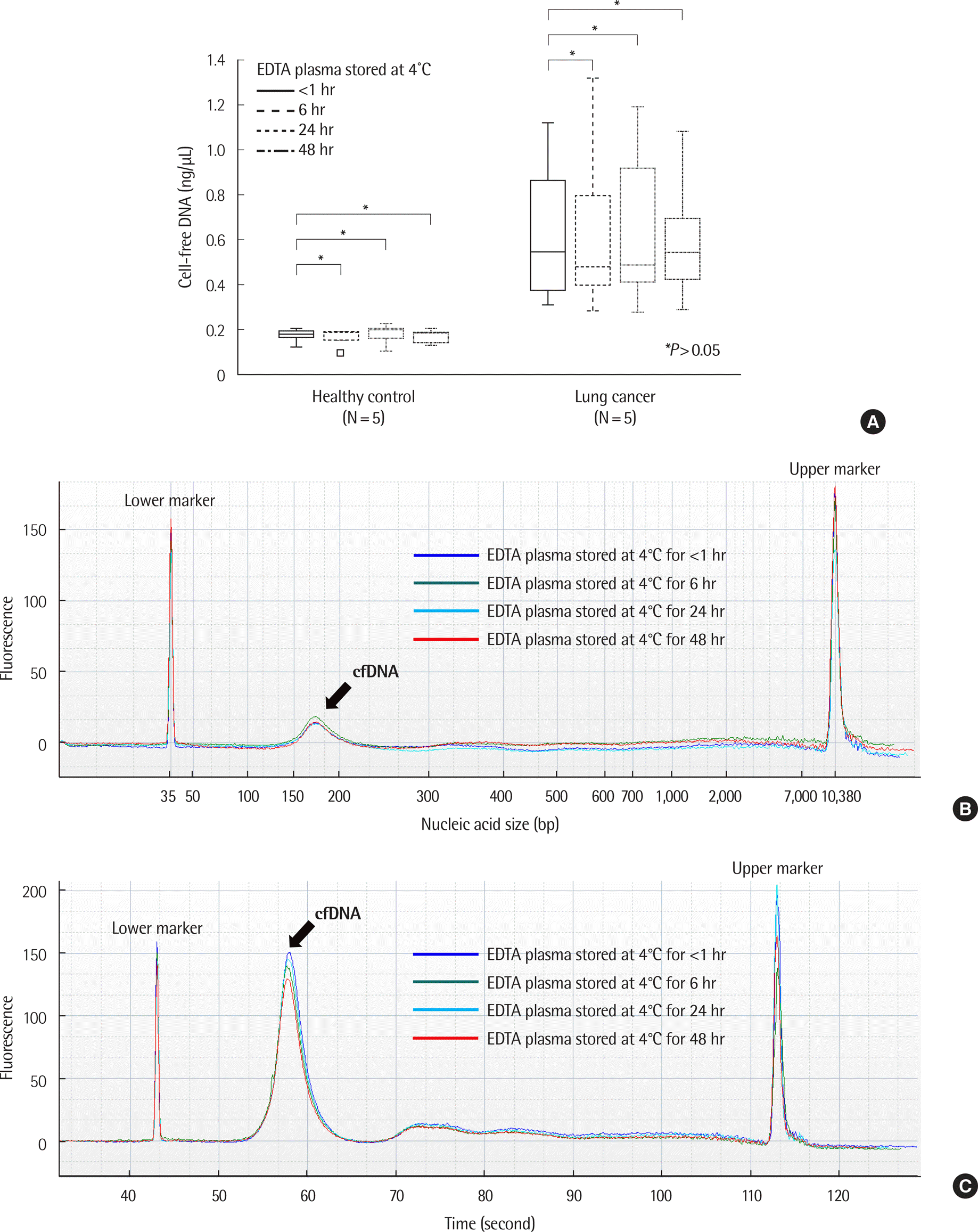
Fig. 4.
Comparison of cell-free DNA (cfDNA) in EDTA containers depending on time of centrifugation. X-axis represents centrifugation condition (A, immediate: immediate plasma separation from EDTA whole blood and the plasma stored at 4˚C; B, delayed: stored EDTA whole blood at 4˚C and delayed plasma separation). (A) The concentration of cfDNA increased when EDTA whole blood was stored at 4˚C for more than 24 hr. (B) Example of size distribution of nucleic acids in EDTA plasma over time in healthy control (case number: HC13). No significant differences were observed according to storage time. (C) Example of size distribution of nucleic acids in EDTA whole blood over time in healthy control (case number: HC13). Large-sized nucleic acids were observed with increasing storage time.
Table 1.
Clinical characteristics of five patients with lung cancer
| Case No. | Age at diagnosis | Sex | Histology of lung cancer | EGFR mutation analysis (date) | |
|---|---|---|---|---|---|
| Tissue biopsy∗ | Plasma† | ||||
| LC06 | 59 | F | NSCLC (adenocarcinoma) | L858R (Aug/2016) | L858R (Oct/2017) |
| LC07 | 65 | F | NSCLC (adenocarcinoma) | Exon 19 deletion (Oct/2014) | No mutation detected (Oct/2017) |
| LC08 | 80 | F | NSCLC (adenocarcinoma) | Exon 19 deletion (Oct/2017) | Exon 19 deletion (Oct/2017) |
| LC09 | 69 | F | NSCLC (adenocarcinoma) | Exon 19 deletion, T790M (Mar/2017) | No mutation detected (Oct/2017) |
| LC10 | 58 | F | NSCLC (adenocarcinoma) | Exon 19 deletion (Sep/2015) | Exon 19 deletion (Oct/2017) |
Table 2.
Cell-free DNA yield according to the specimen container and storage duration
Table 3.
Cell-free DNA yield according to centrifugation conditions in K2-EDTA containers
Table 4.
Detection of EGFR gene mutation in plasma samples from patients with lung cancer
| Sample collection and storage conditions | Input plasma volume (mL) | cfDNA extraction kit manufacturer | Case No. LC06 (L858R) | Case No. LC08 (Exon 19 deletion) | Case No. LC10 (Exon 19 deletion) |
|---|---|---|---|---|---|
| EGFR mutation analysis (semi-quantitative index)∗ | |||||
| K2-EDTA, <1 hr | 2 | Roche | L858R (5.70) | Exon 19 deletion (10.97) | Exon 19 deletion (9.42) |
| K2-EDTA, <1 hr | 1 | Thermo Fisher | L858R (5.12) | Exon 19 deletion (8.41) | Exon 19 deletion (8.71) |
| K2-EDTA, 6 hr | 1 | Thermo Fisher | L858R (5.79) | Mutation not detected | Exon 19 deletion (8.23) |
| K2-EDTA, 24 hr | 1 | Thermo Fisher | L858R (5.03) | Exon 19 deletion (10.17) | Exon 19 deletion (8.97) |
| K2-EDTA, 48 hr | 1 | Thermo Fisher | L858R (4.00) | Exon 19 deletion (8.40) | Exon 19 deletion (8.69) |
| BCT, 1 day | 1 | Thermo Fisher | L858R (6.04) | Exon 19 deletion (9.32) | Exon 19 deletion (6.98) |
| BCT, 3 days | 1 | Thermo Fisher | L858R (6.98) | Exon 19 deletion (9.01) | Exon 19 deletion (8.26) |
| BCT, 7 days | 1 | Thermo Fisher | L858R (6.00) | Exon 19 deletion (9.22) | Exon 19 deletion (8.71) |
| BCT, 14 days | 1 | Thermo Fisher | L858R (4.92) | Exon 19 deletion (9.12) | Exon 19 deletion (8.66) |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download