Abstract
Loss of heterozygosity (LOH) in chromosome 6p has been reported in a number of tumors and some hematologic malignancies, including ALL. LOH in chromosome 6p, on which the HLA genes are located, can give rise to false homozygosity results in HLA genotyping of patients with hematologic malignancies. Here we report false homozygosity results in HLA genotyping due to the loss of whole chromosome 6 in the neoplastic cells of a patient with ALL. A 33-yr-old Korean female patient was admitted for the evaluation of leukocytosis detected during a workup for headache. Her initial white blood cell count was 336.9×109/L with 84% of blasts in the differential count. Precursor-B lymphoblastic leukemia was diagnosed from a subsequent bone marrow study. HLA high-resolution genotyping of the patient was requested at the time of diagnosis for possible hematopoietic stem cell transplantation. Homozygosity results (A*02:01, B*54:01, C*08:01, DQB1*04:01) were obtained, except for the DRB1 locus (DRB1*04:05, DRB1*11:01), in sequence-based typing. Conventional karyotyping of bone marrow metaphase cells revealed chromosomal abnormalities, with loss of multiple chromosomes including chromosome 6, and reduplication of the remaining chromosomes: 29,X,+X,+8,inv(9)(p11q13),+10,+14,+18,+21[15]/58,idemX2[3]/46,XX,inv(9)[2]. LOH at the HLA region was suspected and HLA genotyping was repeated with the peripheral blood in remission state after induction chemotherapy. All 5 HLA loci were typed as heterozygous (A*02:01, A*02:06, B*40:01, B*54:01, C*03:04, C*08:01, DRB1*04:05, DRB1*11:01, DQB1*03:01, DQB1*04:01). To avoid false HLA typing results in patients with hematologic malignancies, clinicians, as well as laboratory personnel, need to be aware of such problems and take appropriate precautions.
The MHC is a genomic region located on the short arm of chromosome 6 (6p), encoding HLA molecules. Loss or down-regulation of MHC class I expression in cancer cells is a major immune escape route used by a large variety of human tumors to evade anti-tumor immune responses mediated by cytotoxic T lymphocytes [1-3]. Multiple mechanisms are responsible for such HLA class I alterations [4-11]. Among them, loss of heterozygosity (LOH) of chromosome 6p has been reported in a number of solid tumors and some hematologic malignancies, mainly in ALL [12, 13].
In patients with leukemia, LOH at the MHC region can give false homozygosity results in HLA typing when the typing is performed using peripheral blood containing a high proportion of leukemic cells. However, such a problem has rarely been reported because HLA typing is not usually carried out on a blood specimen containing a high proportion of neoplastic cells. A case of a CLL patient has been reported involving LOH at the MHC region at the initial HLA typing, which was then detected at confirmatory typing for hematopoietic stem cell transplantation [14]. Here we report false homozygosity results in HLA genotyping due to LOH associated with the loss of whole chromosome 6 in an adult Korean patient with ALL.
A 33-yr-old Korean female was admitted to the hemato-oncology ward of Seoul National University Hospital on October 4, 2010 for the evaluation of leukocytosis found during a workup for headache. A complete blood cell count showed a hemoglobin level of 7.8 g/dL, a platelet count of 15×109/L, and a white blood cell count of 336.9×109/L, of which 84% were blasts. Bone marrow aspirates showed hypercellularity with 99.6% of blasts. The blasts were homogeneously small sized with high nuclear/cytoplasmic ratio and inconspicuous nucleoli. Flow cytometric analysis revealed that the neoplastic cells were positive for TdT, CD19, CD34, and cytoplasmic CD79a, and a diagnosis of precursor-B ALL was made.
On October 7, 2010 (the third day of hospitalization), before initiation of induction chemotherapy, high-resolution HLA DNA typing of the patient was performed using the peripheral blood sample. For future hematopoietic stem cell transplantation, HLA class I (A, B, and C) serological typing of her 2 siblings was also carried out. High-resolution HLA typing was performed by sequence-based typing (SBT) using the AlleleSEQR® HLA Sequencing kit (Atria Genetics, South San Francisco, CA, USA). HLA genotyping of the patient revealed homozygosity at class I and II loci, except for the DRB1 locus: A*02:01, B*54:01, C*08:01, DRB1*04:05, DRB1*11:01, DQB1*04:01 (Table 1, Fig. 1). One of the deduced HLA haplotypes (A*02:01-B*54:01-C*08:01-DRB1*11:01-DQB1*04:01) of the patient was a fairly unusual one for Korean ethnicity. Moreover, homozygosity at the HLA-B locus (B*54:01) in the patient could not be explained, considering the HLA-B types of her 2 siblings (B51, B54 in one and B60, B62 in the other). Retyping of the patient's DNA for HLA-A, -B, and -DR loci by the sequence specific oligonucleotide (SSO) method using Dynal RELI™ SSO HLA-B, -DRB, and -DQB1 kits (Invitrogen, Wirral, UK) also revealed homozygosity at the following loci: HLA-B*54:01 group, DRB1*04:05 group (DRB4*01:01 group), and DQB1*04:01 group. A retrospective review revealed that the patient's HLA genotyping had been requested when she had a high blast count in the peripheral blood, with a white blood cell count of 91.7×109/L, of which 87% were blasts.
Conventional karyotyping performed on a bone marrow aspirate sample obtained on October 11, 2010 (the seventh day of hospitalization) revealed chromosomal abnormalities with loss of multiple chromosomes, including chromosome 6, and reduplication of the remaining chromosomes: 29,X,+X,+8,inv(9)(p11q13),+10,+14,+18,+21[15]/58,idemX2[3]/46,XX,inv(9)[2] (Fig. 2). Retrospective analysis of short tandem repeat (STR) markers revealed LOH in the STR markers on chromosomes 3, 4, 5, 7, 12, and 13 (Table 2), which was in accordance with conventional karyotyping results. Minor alleles due to loss of pertinent chromosomes in neoplastic cells comprised on average 4.1% (range 2.9-6.9%).
False homozygosity results due to LOH at the HLA region were suspected. HLA genotyping was repeated on December 13, 2010, with the peripheral blood in a remission state, 67 days after the initiation of induction chemotherapy. A complete blood cell count showed a hemoglobin level of 10.0 g/dL, a platelet count of 160×109/L, and a white blood cell count of 2.3×109/L, of which 0% were blasts. All of the HLA-A, -B, -C, -DRB1, and -DQB1 loci were typed as heterozygous: A*02:01, A*02:06, B*40:01, B*54:01, C*03:04, C*08:01, DRB1*04:05, DRB1*11:01, DQB1*03:01, DQB1*04:01 (Table 1, Fig. 1). Karyotyping performed on a peripheral blood sample obtained on the same date (in a remission state) revealed a normal karyotype: 46,XX,inv(9)(p11q13)[20]. STR analysis performed on the same blood sample also revealed heterozygosity results in all 9 markers. Thus the heterozygosity results of HLA-A, -B, -C, -DR, and -DQ loci were confirmed and finally used to search for an unrelated donor for bone marrow transplantation.
HLA class I antigens are vital in the recognition of tumor cells by tumor-specific cytotoxic T lymphocytes. Loss or down-regulation of HLA class I antigens, therefore, represents a way by which tumors can escape T-cell surveillance, adversely affecting the course of disease and the outcome of T-cell-based immunotherapy [1-3]. This can have several causes: absence of β2-microglobulin or transporter associated with antigen processing (TAP) expression [4-6]; loss of heterozygosity, large deletions, or mitotic recombination in chromosome 6 [4, 8, 9]; transcriptional down-regulation [10]; or point mutation, partial deletion, or somatic recombination [4, 7, 11].
LOH in chromosome 6p has been detected in about 10-55% of solid tumors such as those of the colon or larynx, melanoma, and breast cancer [12, 15]. Previously reported frequencies of LOH in 6p in hematologic malignancies are variable according to the detection method and definition of LOH. Masuda et al. reported loss or down-regulation of HLA class I expression in only 4 out of 43 freshly isolated adult hematologic malignancy samples and no haplotype loss in the HLA genome [16]. In childhood ALL, the frequency of LOH in 6p has been reported to be 7-8% [17, 18]. In contrast, McEvoy et al. reported a high frequency of MHC LOH in ALL patients (19/56), which was usually associated with whole chromosome 6 loss (13/19) [13].
LOH in chromosome 6p in hematologic malignancies can cause false homozygosity results in HLA genotyping when the typing is performed with leukemic blood samples. This can give rise to a serious problem in hematopoietic stem cell transplantation in that the patient would be at a risk of being transplanted from a falsely matched donor if the false typing results are not properly recognized. Serologic typing of HLA antigens with blood samples containing high blast counts is difficult, and therefore, typing is usually not attempted. Nowadays DNA typing is increasingly used for HLA typing, and such problems may occur if clinicians inappropriately order HLA genotyping with leukemic blood samples, and the HLA typing laboratory is not aware of the problem in the blood sample requested for typing. False homozygosity results for HLA class I due to LOH at the MHC region has been reported in a CLL patient, which was detected at confirmatory typing for hematopoietic stem cell transplantation [14]. In this patient, LOH in chromosome 6p was confirmed by microarray-based comparative genomic hybridization of tumor cells.
In our case, whole chromosome 6 loss in leukemic cells caused false homozygosity results in HLA-A, -B, -C, and -DQB1 SBT and HLA-B, -DRB, and -DQB1 SSO typing. Retesting with a peripheral blood sample in a remission state using SBT revealed heterozygosity results in all of the 5 HLA loci tested. In HLA-DRB1 typing in the initial sample, a heterozygosity result was obtained by SBT and homozygosity result by SSO typing. A sequencing electropherogram showed amplification peaks of the minor allele (DRB1*11:01, present only in a small fraction of normal leukocytes) which were much lower than those of the major allele (DRB1*04:05, present in both leukemic blasts and normal leukocytes) (Fig. 1). As for the discrepancy in HLA-DRB1 typing results between the 2 typing methods, differences in the primers used for these methods might be the cause. For HLA-DRB1-specific amplification, different HLA-DRB1 alleles cannot be amplified by a single pair of primers and thus several different primer pairs amplifying different groups of DRB1 alleles are used in HLA-DRB1 SBT. The primer pairs that can amplify each of the 2 DRB1 alleles (DRB1*04:05, *11:01) in this patient are expected to be different and the minor allele could thus be amplified in SBT. In HLA-DRB SSO typing, using a Dynal RELI™ SSO kit, single primer pairs amplifying DRB1/DRB3/DRB4/DRB5 genes are used, and the minor allele in this patient seems not to have been effectively amplified to be detected. Likewise, minor alleles in HLA-A, -B, -C, and -DQB1 loci in this patient seem not to have been effectively amplified to be detected by SBT. Thus, when HLA typing is performed on blood samples containing a high proportion of blasts with LOH in chromosome 6p, typing results may differ between different typing methods according to differences in their detection sensitivity. In the PCR-sequence specific primer (SSP) method, different primer pairs amplify different HLA alleles, and this method is expected to detect minor alleles with higher sensitivity than the SSO or SBT methods. We later performed PCR-SSP typing (AllSet Gold SSP HLA typing kit, Invitrogen, Brown Deer, WI, USA) for HLA-B (48 primer pairs) and -DR loci (24 primer pairs) using the DNA sample from our patient at the time of diagnosis. As expected, the PCR-SSP method detected heterozygosity for both HLA-B and -DR loci, although the minor alleles showed much weaker amplification bands on the electropherogram than the major alleles.
To avoid false HLA typing results in patients with hematologic malignancies, clinicians as well as laboratory personnel need to be aware of such problems and take appropriate precautions. In leukemia patients, HLA typing is usually performed at the time of diagnosis or during the first complete remission [19]. Clinicians should not request HLA typing using blood samples at the time of diagnosis if patients have blasts in the peripheral blood. There is no data available on what percentage of blasts or neoplastic cells in the white cell differential count of a peripheral blood sample can be present without affecting the HLA molecular typing results in patients with LOH in 6p. Because blasts generally have considerably larger nuclei than normal leukocytes, the DNA fraction from blasts would comprise a much larger portion of total DNA extracted from leukocytes than that simply calculated from the percentage of blasts in the blood sample. This is evidenced in our case; whereas the blood sample used for HLA DNA typing contained 87% blasts, the minor alleles detected by STR analysis comprised a much lower percentage (average 4.1%) than expected (above 10%). If HLA typing is required in such patients before remission is obtained, alternative samples, such as a buccal swab or hair follicles, should be considered for HLA typing. HLA typing laboratories involved in the typing of patients with hematologic malignancies should be aware of such problems and should check patients' hematologic data whenever they have unusual HLA typing results or discrepancies between different typing methods. In addition, the patients' HLA typing results should be reconfirmed by confirmatory typing. For hematopoietic stem cell transplantation, confirmatory HLA typing using newly collected samples should be performed not only for donors but also for patients [20, 21]. However, confirmatory HLA typing of patients is not reimbursed by the national medical insurance system and is not routinely carried out as a standard clinical practice in this country.
We have described a case of false homozygosity results in HLA genotyping in a patient with precursor-B ALL due to the loss of whole chromosome 6 in the neoplastic cells. To avoid false HLA typing results in patients with hematologic malignancies, clinicians and HLA typing laboratory personnel need to be aware of the problem and take appropriate precautions. HLA typing should not be performed with blood samples containing a high proportion of neoplastic cells, and alternative samples of buccal swab or hair follicles should be used if needed. In addition, confirmatory HLA typing should be routinely performed for hematopoietic stem cell transplantation.
References
1. Garrido F, Cabrera T, Concha A, Glew S, Ruiz-Cabello F, Stern PL. Natural history of HLA expression during tumour development. Immunol Today. 1993; 14:491–499. PMID: 8274189.

2. Ferrone S, Marincola FM. Loss of HLA class I antigens by melanoma cells" molecular mechanisms, functional significance and clinical relevance. Immunol Today. 1995; 16:487–494. PMID: 7576053.
3. Garrido F, Ruiz-Cabello F, Cabrera T, Pérez-Villar JJ, López-Botet M, Duggan-Keen M, et al. Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol Today. 1997; 18:89–95. PMID: 9057360.

4. Browning M, Petronzelli F, Bicknell D, Krausa P, Rowan A, Tonks S, et al. Mechanisms of loss of HLA class I expression on colorectal tumor cells. Tissue Antigens. 1996; 47:364–371. PMID: 8795136.

5. D'Urso CM, Wang ZG, Cao Y, Tatake R, Zeff RA, Ferrone S. Lack of HLA class I antigen expression by cultured melanoma cells FO-1 due to a defect in B2m gene expression. J Clin Invest. 1991; 87:284–292. PMID: 1898655.
6. Spies T, DeMars R. Restored expression of major histocompatibility class I molecules by gene transfer of a putative peptide transporter. Nature. 1991; 351:323–324. PMID: 2034277.

7. Browning MJ, Krausa P, Rowan A, Hill AB, Bicknell DC, Bodmer JG, et al. Loss of human leukocyte antigen expression on colorectal tumor cell lines: implications for anti-tumor immunity and immunotherapy. J Immunother Emphasis Tumor Immunol. 1993; 14:163–168. PMID: 8297898.
8. Koopman LA, Mulder A, Corver WE, Anholts JD, Giphart MJ, Claas FH, et al. HLA class I phenotype and genotype alterations in cervical carcinomas and derivative cell lines. Tissue Antigens. 1998; 51:623–636. PMID: 9694355.

9. Torres MJ, Ruiz-Cabello F, Skoudy A, Berrozpe G, Jimenez P, Serrano A, et al. Loss of an HLA haplotype in pancreas cancer tissue and its corresponding tumor derived cell line. Tissue Antigens. 1996; 47:372–381. PMID: 8795137.

10. Soong TW, Hui KM. Locus-specific transcriptional control of HLA genes. J Immunol. 1992; 149:2008–2020. PMID: 1517566.
11. Koopman LA, van Der Slik AR, Giphart MJ, Fleuren GJ. Human leukocyte antigen class I gene mutations in cervical cancer. J Natl Cancer Inst. 1999; 91:1669–1677. PMID: 10511595.

12. Jiménez P, Cantón J, Collado A, Cabrera T, Serrano A, Real LM, et al. Chromosome loss is the most frequent mechanism contributing to HLA haplotype loss in human tumors. Int J Cancer. 1999; 83:91–97. PMID: 10449614.
13. McEvoy CR, Morley AA, Firgaira FA. Evidence for whole chromosome 6 loss and duplication of the remaining chromosome in acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2003; 37:321–325. PMID: 12759931.

14. Coppage M, Busacco A, Lenog N, Rothberg P, Phillips G, Becker M. Leukemia specific loss of heterozygosity at MHC locus detected at confirmatory typing of HSCT recipient with CLL. Hum Immunol. 2010; 71(S):S130.
15. Maleno I, López-Nevot MA, Cabrera T, Salinero J, Garrido F. Multiple mechanisms generate HLA class I altered phenotypes in laryngeal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Cancer Immunol Immunother. 2002; 51:389–396. PMID: 12192539.

16. Masuda K, Hiraki A, Fujii N, Watanabe T, Tanaka M, Matsue K, et al. Loss or down-regulation of HLA class I expression at the allelic level in freshly isolated leukemic blasts. Cancer Sci. 2007; 98:102–108. PMID: 17083564.

17. Baccichet A, Qualman SK, Sinnett D. Allelic loss in childhood acute lymphoblastic leukemia. Leuk Res. 1997; 21:817–823. PMID: 9393596.

18. Takeuchi S, Bartram CR, Wada M, Reiter A, Hatta Y, Seriu T, et al. Allelotype analysis of childhood acute lymphoblastic leukemia. Cancer Res. 1995; 55:5377–5382. PMID: 7585604.
19. Ljungman P, de Witte T, Verdonck L, Gahrton G, Freycon F, Gravett P, et al. Bone marrow transplantation for acute myeloblastic leukaemia: an EBMT Leukaemia Working Party prospective analysis from HLA-typing. Br J Haematol. 1993; 84:61–66. PMID: 8338779.

20. . Standards for histocompatibility testing, version 5.6.1. Updated on Oct 2009. http://www.efiweb.eu/index.php?id=102.
21. Treleaven JG, Barrett AJ, editors. Hematopoietic stem cell transplantation in clinical practice. 2008. 1st ed. Churchill Livingstone;p. 224.
Fig. 1
Sequence-based typing of HLA-B and -DRB1 genes at the time of diagnosis (A, B) and at remission (C, D). HLA alleles detected were: (A) B*54:01; (B) DRB1*04:05, *11:01; (C) B*40:01, *54:01; (D) DRB1*04:05, *11:01. At the time of diagnosis, the minor HLA-B allele (B*40:01) shows amplification peaks indistinguishable from background noise (A), whereas the minor HLA-DRB1 allele (DRB1*11:01) shows lower but discernible amplification peaks (B). At remission, both HLA-B and -DRB1 loci show heterozygosity with similar amplification peaks for the 2 alleles. E2F and E2R represent exon 2 forward and exon 2 reverse, respectively.
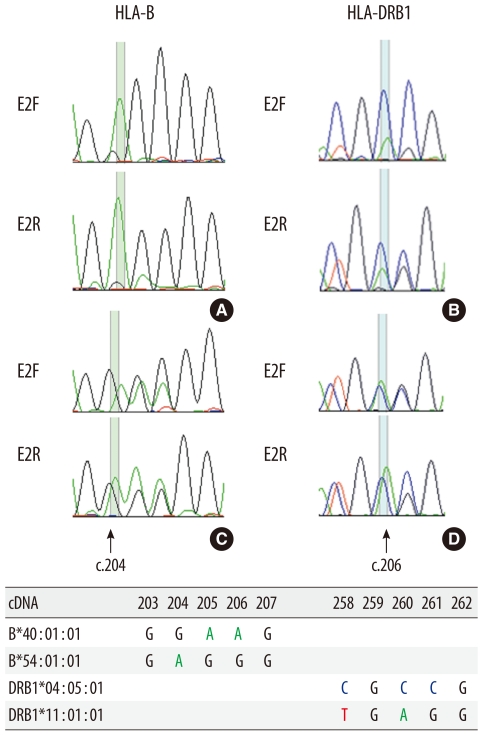
Fig. 2
Initial karyotyping of G-banded bone marrow metaphase cells showing loss of multiple chromosomes including chromosome 6 (A) and reduplication of the remaining chromosomes (B).
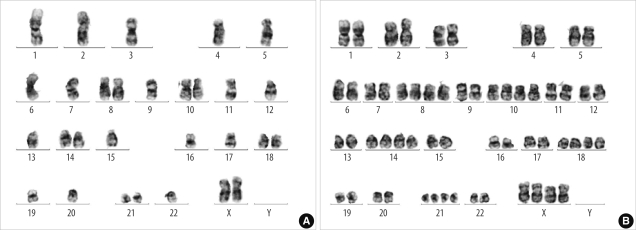
Table 1
HLA sequence-based typing results of the patient using blood samples obtained at the time of diagnosis and in a remission state, and serotyping results of her 2 siblings
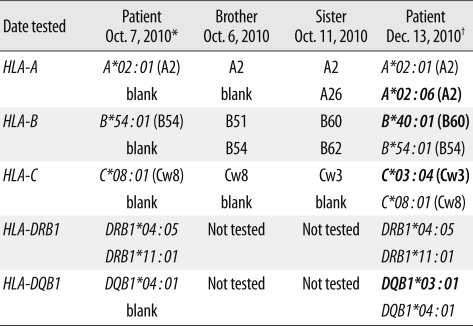
Table 2
Results of the short tandem repeat (STR) analysis of 9 loci using patient's DNA at the time of diagnosis and in a remission state
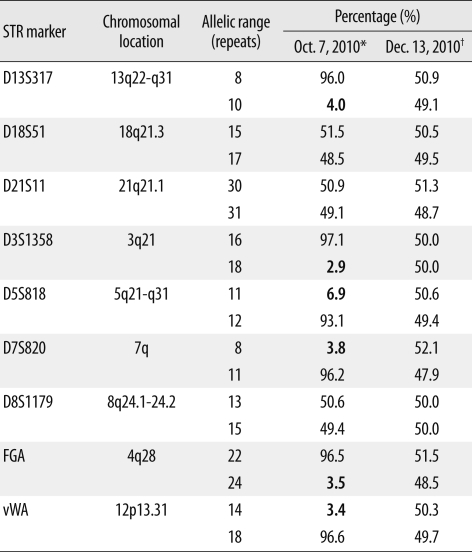
*Initial blood sample (87% blasts) obtained before induction chemotherapy. Minor alleles (average 4.1%, range 2.9-6.9%) due to loss of pertinent chromosomes in neoplastic cells (Fig. 2) are in bold; †Blood sample (0% blasts) obtained in a remission state after chemotherapy.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download