Abstract
Background
Leukapheresis has commonly been used to obtain the cell products intended for clinical cell therapy. Hypocalcemia related to citrate toxicity and some circulatory effects such as hypovolemia and hypotension are well-known complications of leukapheresis. In this study, we analyzed the gene expression profiles of peripheral blood mononuclear cells (PBMCs) obtained before and after leukapheresis to determine if the hemodynamic changes can affect the gene expression profiles of leukocytes.
Methods
PBMCs were isolated from EDTA blood from 5 healthy donors collected before and immediately after apheresis. RNA was isolated, amplified, and analyzed using a cDNA microarray with 17,500 genes. Hierarchical clustering analysis was performed to evaluate the differences of gene expression profiling.
Results
Hierarchical clustering separated PBMCs from different donors with each other, but did not separate PBMCs collected before and after leukapheresis. Comparison of gene expression by PBMCs collected before and after leukapheresis found only 25 genes were differentially expressed (15 were up-regulated and 10 were down-regulated after leukapheresis) (F-test, P<0.005). Stress induced apoptosis-related genes, ANXA3, DEDD, and ATXN2L, and cytokine-related genes, IL13RA1 and IK, which were also related to stress, were up-regulated after leukapheresis. Genes involved in DNA and protein binding, such as CLSTN3, LRBA, SATB2, and HSPA8, were down-regulated.
REFERENCES
1.Hashimoto SI., Suzuki T., Nagai S., Yamashita T., Toyoda N., Matsushima K. Identification of genes specifically expressed in human activated and mature dendritic cells through serial analysis of gene expression. Blood. 2000. 96:2206–14.

2.Dauer M., Obermaier B., Herten J., Haerle C., Pohl K., Rothenfusser S, et al. Mature dendritic cells derived from human monocytes within 48 hours: a novel strategy for dendritic cell differentiation from blood precursors. J Immunol. 2003. 170:4069–76.

3.Sallusto F., Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994. 194:1109–18.

4.Felzmann T., Witt V., Wimmer D., Ressmann G., Wagner D., Paul P, et al. Monocyte enrichment from leukapharesis products for the generation of DCs by plastic adherence, or by positive or negative selection. Cytotherapy. 2003. 5:391–8.

5.Han KS, Park MH, editors. Transfusion medicine. 2nd ed.Seoul: Korea Medical Book Publisher Co;1999. p. 10. (한규섭, 박명희, 등. 수혈의학. 제2판. 서울: 고려의학 1999 10).

6.Brecher ME, editor. Technical manual. 15th ed.Bethesda: American Association of Blood Banks;2005. p. 587.
7.Ottinger HD., Beelen DW., Scheulen B., Schaefer UW., Grosse-Wilde H. Improved immune reconstitution after allotransplantation of peripheral blood stem cells instead of bone marrow. Blood. 1996. 88:2775–9.

8.Whang DH., Hur M., Park KU., Shin S., Kim YH., Shin HY, et al. Experience with 671 peripheral blood stem cell collection. Korean J Blood Transfus. 2000. 11:145–56. (황동희, 허미나, 박경운, 신수, 김양현, 신희영등. 말초혈액조혈모세포채집술 671예시행경험. 대한수혈학회지 2000;11: 145-56.).
9.Wang E., Miller LD., Ohnmacht GA., Liu ET., Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000. 18:457–9.

10.Wang E., Miller LD., Ohnmacht GA., Mocellin S., Perez-Diez A., Petersen D, et al. Prospective molecular profiling of melanoma metastases suggests classifiers of immune responsiveness. Cancer Res. 2002. 62:3581–6.
11.Eisen MB., Spellman PT., Brown PO., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998. 95:14863–8.

12.Han J., Farnsworth RL., Tiwari JL., Tian J., Lee H., Ikonomi P, et al. Quality prediction of cell substrate using gene expression profiling. Genomics. 2006. 87:552–9.

13.Park BJ., Kim GK. Gene expression profiling of meningioma by cDNA chip. J Korean Neurosurg Soc. 2004. 35:560–8. (박봉진 및 김국기. 수막종에서cDNA chip을이용한유전자발현연구. J Korean Neurosurg Soc 2004;35: 560-8.).
14.Baechler EC., Batliwalla FM., Karypis G., Gaffney PM., Moser K., Ortmann WA, et al. Expression levels for many genes in human peripheral blood cells are highly sensitive to ex vivo incubation. Genes Immun. 2004. 5:347–53.

15.Ng H., Pulst SM., Huynh DP. Ataxin-2 mediated cell death is dependent on domains downstream of the polyQ repeat. Exp Neurol. 2007. 208:207–15.

16.Wiedemeyer R., Westermann F., Wittke I., Nowock J., Schwab M. Ataxin-2 promotes apoptosis of human neuroblastoma cells. Oncogene. 2003. 22:401–11.

17.Kim YM., Kim HJ., Song EJ., Lee KJ. Glucuronic acid is a novel inducer of heat shock response. Mol Cell Biochem. 2004. 259:23–33.
Fig. 1.
Gene expression profiles of PBMCs obtained before and after leukapheresis. The 8,661 genes that remained after filtering (expressed in 80% of samples) were analyzed. Hierarchical clustering separated PBMCs from different donors with each other, but did not separate PBMCs collected before and after leukapheresis. Pre-PBMC, PBMCs obtained before leukapheresis; Post-PBMC, PBMCs obtained after leukapheresis.
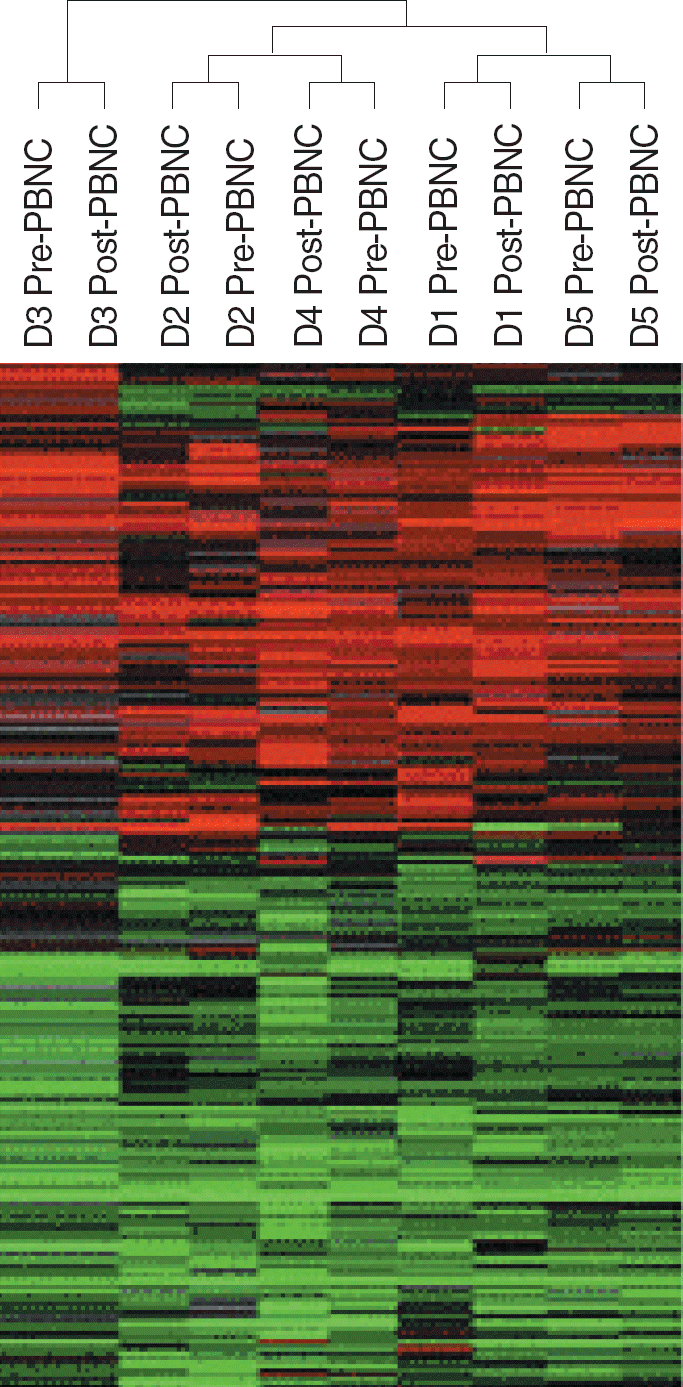
Table 1.
Characteristics of donors and samples
| Donor No | Age | Sex | Race | Cell separator | Product volume | Cell counts of products | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hb (g/dL) | WBC (× 103/μL) | PLT (× 103/L) | |||||||||
| pre∗ | post† | pre | post | pre | post | ||||||
| 1 | 48 | F | African-American | Cobe Spectra | 136 mL | 11.9 | 11.0 | 4.8 | 4.5 | 275 | 174 |
| 2 | 68 | M | Caucasian | Cobe Spectra | 128 mL | 13.6 | 13.6 | 5.4 | 5.7 | 196 | 135 |
| 3 | 32 | M | African-American | CS3000 | 136 mL | 14.5 | 14.1 | 3.8 | 3.4 | 244 | 176 |
| 4 | 29 | M | Hispanic | Cobe Spectra | 137 mL | 14.3 | 14.6 | 6.1 | 5.7 | 281 | 218 |
| 5 | 56 | M | Caucasian | Cobe Spectra | 150 mL | 15.0 | 15.0 | 4.5 | 4.4 | 239 | 156 |
Table 2.
Genes differentially expressed by post-leukapheresis PBMCs compared to pre-leukapheresis PBMCs




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download