Abstract
Background
Human papillomavirus (HPV) infection causes cervical cancer, thus necessitating early detection by screening. Rapid and accurate HPV genotyping is crucial both for the assessment of patients with HPV infection and for surveillance studies.
Methods
Fifty-eight cervicovaginal samples were tested for HPV genotypes using four methods in parallel: nested-PCR followed by conventional sequencing, INNO-LiPA, electrochemical DNA chip, and next-generation sequencing (NGS).
Human papillomavirus (HPV) infection is one of the most common causes of sexually transmitted disease [1]. Young women are more likely than men to become infected with HPV and often contract multiple strains of the virus [2]. To date, over 120 different types of HPV have been identified, approximately 40 of which can infect the cervicovaginal mucosa. High-risk HPV (HR-HPV) types are strongly associated with premalignant and malignant cervical lesions, while low-risk HPV (LR-HPV) types are primarily linked to benign anogenital warts [34].
HPV has a circular double-stranded DNA genome of approximately 8,000 bp. The genome is divided into early (E), late (L), and noncoding (upstream regulatory) regions. Variations among HPV genotypes occur primarily in the L1 region, where nucleotide sequence variation may be greater than 10% [4].
There are several molecular diagnostic tests for detecting HPV DNA including direct sequencing, hybridization with genotype-specific probes, or restriction fragment length polymorphism analysis [56]. Although a combination of several techniques may be used, identification of HPV DNA has traditionally relied on PCR amplification of the major capsid gene, L1, using the degenerate primers, MY09/11 [7]. Alternatively, primers GP5+/GP6+ (extended versions of the MY primers) have been used to amplify a 140–150 bp fragment of the L1 region, resulting in higher detection sensitivity [8]. This combination of primers gives rise to increased PCR accuracy, although the assay may not be reliable for detecting multiple HPV genotypes [9].
The INNO-LiPA HPV Assay (Innogenetics, Gent, Belgium) is a commercially available HPV genotyping test based on reverse hybridization of amplified HPV products onto a membrane strip containing multiple probes immobilized as parallel lines, which can detect 28 different HPV genotypes [1011]. Alternatively, the electrochemical DNA chip system can detect single or multiple infections caused by 13 HR-HPV genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68); all processes of this system, from reaction to measurement and analysis, are integrated into a single compact piece of equipment, which can determine HPV genotype relatively quickly [12].
The emergence of next-generation sequencing (NGS) technologies provides an opportunity to directly examine viral diversity in clinical samples without previous sequence information [13]. This system can yield data outputs ranging from 300 kilobases to 1 terabase in a single run and uses primers tagged with initial sequences of specific nucleotides to identify each sample [14]. This results in a massive number of parallel sequencing reactions and DNA fragments, all of which possess initial sequence tags specific to the original individual samples (barcodes), which are analyzed molecule by molecule. This method is not widely used for HPV diagnosis, and its accuracy is yet to be confirmed [15]. The objective of this study was to compare the diagnostic performance of four techniques for HPV detection and genotyping: nested-PCR followed by conventional sequencing, INNO-LiPA, electrochemical DNA chip, and NGS.
This study was approved by the Institutional Review Board (IRB) of the Faculty of Medicine, Chulalongkorn University (IRB number 603/2558). The IRB waived the need for consent because the clinical samples were de-identified and anonymous. All experiments in this research involved conveniently archived samples derived from an earlier study [16], which examined the prevalence of cervicovaginal HPV infection. Gynecological samples were selected from the samples obtained during routine Pap smear checkups, investigations, or treatment of patients. All cervical cytology was confirmed by a cytotechnologist and a pathologist. Samples were categorized into four groups based on cytology: normal, low-grade squamous intraepithelial lesions (LSIL), high-grade squamous intraepithelial lesions (HSIL), and cervical cancer. The coding of all samples was anonymous. All samples were kept in LBC buffer (ThinPrep; Hologic, West Sussex, UK) or phosphate buffered saline (PBS). Approximately 15 mL of each LBC was centrifuged at 3,000 rpm for 5 minutes, and the supernatant was removed. Then, 800 µL of the sample was added to a 1.5 mL tube to wash the pellet, the sample was centrifuged at 8,000 rpm for 5 minutes, and the supernatant was removed.
DNA was extracted from cervicovaginal samples in LBC using the QIAamp DNA mini kit (QIAGEN, Valencia, CA, USA), according to the manufacturer's protocol. The DNA samples were stored at –20℃ until tested. All samples were tested in parallel for the β-globin gene as an internal control.
DNA was amplified using the nested MY/GP primer set targeting the L1 gene, as previously described [18]. Sanger sequencing was performed by FirstBASE Laboratories SDN BHD (Selangor Darul Ehsan, Malaysia). Nucleotide sequences were analyzed using BLAST via the NCBI website for comparison with HPV sequences in the GenBank database.
HPV genotyping was performed using the INNO-LiPA HPV genotyping Extra test (Innogenetics N.V., Ghent, Belgium), following the manufacturer's instructions. This assay can identify 28 different HPV genotypes, including all known HR-HPV genotypes and probable HR-HPV genotypes (16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82), as well as several LR-HPV genotypes (6, 11, 40, 43, 44, 54, and 70) and a number of additional types (69, 71, and 74), based on nested-PCR amplification of a fragment of the L1 region of the HPV genome. Amplified products were denatured under alkaline conditions and immediately incubated with the test strip in hybridization buffer. The results were visually interpreted by two independent investigators by comparing them with a template provided with the assay.
The electrochemical DNA chip is comprised of specific DNA probes targeting the L1 region of 13 carcinogenic HR-HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68). The assay consists of six loop-mediated isothermal amplification (LAMP) reagents, an intercalation reagent, and an electrochemical DNA chip. The conditions for the LAMP procedure are denaturation at 95℃ for 5 minutes, followed by 65℃ for 90 minutes and 80℃ for 5 minutes. The automated hybridization of probe and primer generates electrochemical signals quantifiable with a GLH-2C601 Genelyzer (Toshiba, Tokyo, Japan). A previous study using the same electrochemical DNA chip demonstrated that there was no cross-hybridization [12].
Fifty-eight cervicovaginal samples (seven normal, 18 LSIL, 12 HSIL, and 21 cervical cancer samples) previously genotyped by nested-PCR, INNO-LiPA, and electrochemical DNA chip, were selected for HPV genotyping by NGS using the MY/GP primer set for amplification. These nested-PCR primers amplify a 180-bp fragment of the L1 region [17]. Amplicons were purified using Expin Gel SV (GeneAll Biotechnology Co., LTD., Seoul, Korea), according to the manufacturer's instructions. Purified DNA concentration was determined using a Qubit fluorometer (Life Technologies Corporation, Carlsbad, CA, USA). DNA libraries were prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina, following the manufacturer's recommendations (New England BioLabs, Herts, UK). Briefly, purified amplicons were end-repaired, adaptor-ligated, and cleaned up. Subsequently, DNA libraries obtained from each sample were amplified by nested-PCR with different index primers and then purified using AmPure XP beads (Beckman Coulter, Porterville, CA, USA). DNA libraries with different index primers were pooled in equal amounts to generate a 2-nM master DNA library, which was denatured with NaOH and then diluted using HT1 buffer to yield a 15 pM DNA library. Paired-end (150×2) deep sequencing was performed using the MiSeq v2 reagent kit (Illumina, Inc., San Diego, CA, USA) on the MiSeq platform (Illumina, Inc.) using a standard protocol.
FASTQ data were processed and analyzed using CLC genomic workbench version 8 (http://www.clcbio.com/). Low-quality reads (Q-score<30) and adaptor sequences were excluded, and low-quality sequence regions were trimmed. The pass-filter reads (Q-score≥30) were aligned with sequences of multiple HPV genotypes obtained from the NCBI and Papillomavirus Episteme (PaVE; http://pave.niaid.nih.gov) databases. HPV genotypes were identified and quantified based on the number of reads matched to each genotype.
The Statistical Package for the Social Sciences (SPSS) software version 17.0 (IBM, Somers, NY, USA) was used for statistical analysis. The Pearson's chi-square test was calculated to compare the detection of HPV genotyping between the four techniques. P values<0.05 were considered statistically significant.
HPV detection results varied substantially between the four methods tested using the same 58 samples. For NGS, DNA samples were combined into three pools of 24, 24, and 10, yielding total sequence outputs of 2.38, 2.44, and 1.64 Gb, respectively. Short sequence reads were trimmed, and a cut-off quality score of 30 was applied. The average number of pass-filter reads per sample was >2×104 (Table 1). Paired-end reads were assembled to generate L1 sequences of approximately 180 bp. These reads were used for HPV genotyping based on sequence comparison with HPV reference sequences.
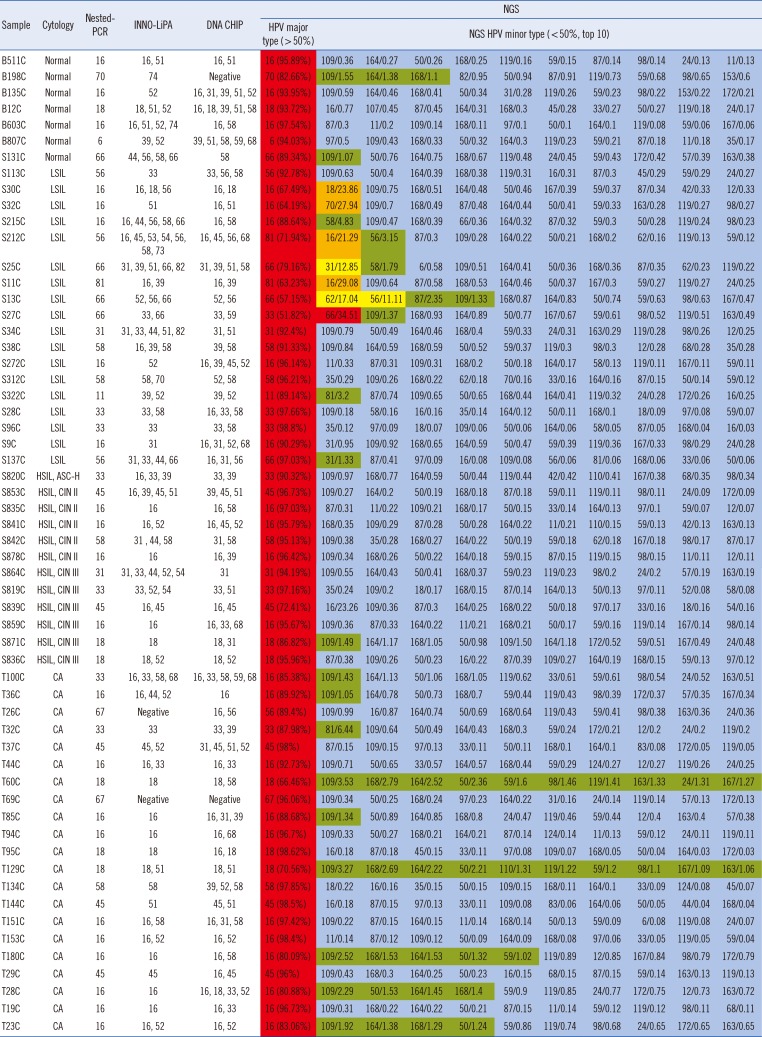
NGS identified a variety of HPV genotypes in each sample; generally, more genotypes were detected using NGS than with the other methods (Table 2). HPV16 was the predominant genotype among the samples analyzed by NGS, consistent with the results of the other methods. Of the sequence reads, >50% matched at least one major HPV genotype. The minor HPV genotypes in each sample (detailed in Table 2) are indicated in red (≥30%), orange (20–29.9%), yellow (10–19.9%), green (1–9.9%), and blue (<1%).
Only a single genotype was identified per sample using the nested-PCR; the most frequent genotype was HPV16 (36.2%). Typing by INNO-LiPA revealed multiple infections in 45 of the 58 samples, with only one sample producing a negative result. The maximum number of genotypes/sample detected by INNO-LiPA was 10. The electrochemical DNA chip analysis findings indicated that 55 samples were from individuals with multiple infections, with a maximum of five genotypes/sample. Multiple infections were identified in all 58 samples by NGS analysis. The Pearson's chi-square test showed a significant difference between NGS and electrochemical DNA chip (P=0.004). In contrast, there was no significant difference between NGS and INNO-LiPA for HPV genotyping. The concordance rate of the four methods (using sequences derived from nested-PCR as gold standard) was 100% for NGS, 77.6% for INNO-LiPA, and 82.8% for electrochemical DNA chip.
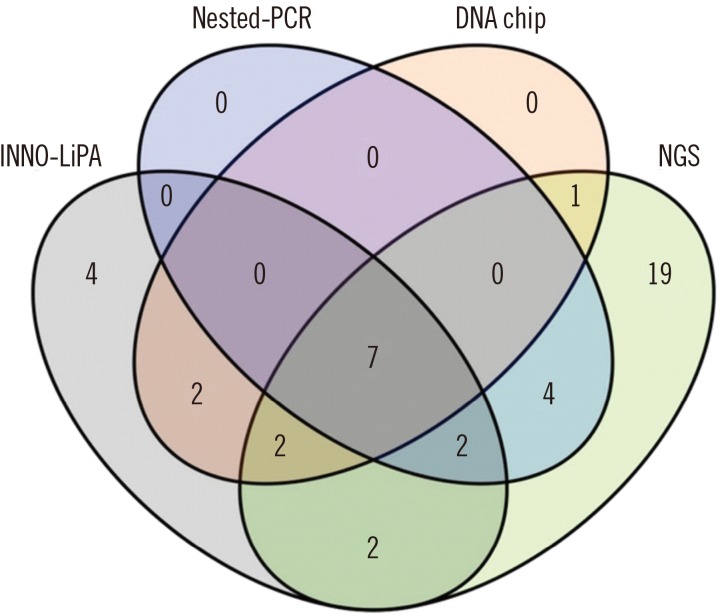
A comparison of the four methods is presented in Fig. 1 as a Venn diagram. Seven HPV genotypes (16, 18, 31, 33, 45, 56, and 58) were identified using all four techniques. All of these were HR-HPV genotypes and were found primarily in samples from malignant or pre-malignant lesions. Interestingly, four genotypes (44, 53, 73, and 74) were detected by INNO-LiPA, but not by the other methods. Nineteen HPV genotypes (12, 24, 42, 50, 57, 62, 87, 97, 98, 109, 110, 119, 124, 153, 163, 164, 167, 168, and 172) identified by NGS were not detected by nested-PCR, INNO-LiPA, or the electrochemical DNA chip method.
HPV genotyping is important for epidemiological studies, potential improvement of the current HPV vaccine, and more effective cervical cancer screening. Currently, several methods are used for HPV genotyping, which differ according to sample preparation, analysis methods, and (for nested-PCR -based assays) primers used. In addition, each method varies in both sensitivity and specificity [18]. Our NGS results were in agreement with those of the other three techniques and were in good agreement with a previous finding, which also showed that NGS had high sensitivity and was suitable for detecting multiple HPV infections [19].
Massively parallel sequencing has transformed genetic research [20], enabling increased characterization of genomes, transcriptomes, epigenomes, and microbiomes [21]. This study presents proof-of-principle that NGS can be used for the characterization of HPV genotypes and suggests its potential advantages over existing hybridization-based genotyping systems. In this study, NGS HPV genotyping was performed using the Illumina MiSeq platform and the genotyping results were compared with those using the other three methods. Comparison of the results demonstrated that 19 HPV genotypes were detected by NGS, but not using the other approaches. These 19 genotypes are not usually identified during HPV screening. HPV genotypes 42, 50, 57, 62, 87, and 97 are associated with LSIL, which causes common skin warts and cutaneous lesions [422232425], while the remaining 13 genotypes detected solely by NGS are largely uncharacterized. While none of these LSIL genotypes confer any measurable increased risk of developing cervical intraepithelial neoplasia, the seven HPV genotypes identified by all four techniques were high-risk genotypes that commonly cause cervical cancer [26].
The results of a previous study using 454 NGS technology and HPV-specific primers to amplify the L1 region demonstrated that the NGS results correlated well with those obtained using the INNO-LiPA HPV Genotyping Extra assay [19]. In this study, the Illumina MiSeq platform HPV typing results were consistent with those generated using the other techniques, indicating that HPV typing using the Illumina MiSeq method is accurate. In contrast, four HPV genotypes were detected by INNO-LiPA, but not robustly identified by NGS. HPV44 was also identified by NGS; however, the frequency of detection was <0.5%. The three other genotypes detected solely by INNO-LiPA, HPV73 (HR-HPV), HPV53 (probably high-risk) [27], and HPV74 (additional genotype), may represent occasional false-positive results generated by this hybridization-based assay. INNO-LiPA utilizes reverse hybridization with multiple probes specific for different genotypes, and interpretation of the results can be quite difficult, depending on the number and density of the bands present on the strip. A number of genotypes, which are not commonly distributed worldwide, may be problematic to interpret.
One limitation of the NGS assay is the amplification of the HPV genome L1 region using the MY/GP primers; these primers yield a 180 bp HPV DNA amplicon, while the INNO-LiPA assay amplifies a similar region using the SPF10 primers, yielding a product of only 65 bp [28]. A previous study demonstrated that the SPF10 primer set is more sensitive than the MY09/11 primers [29]. Moreover, nested-PCR amplicon size can influence assay sensitivity, with smaller amplicons associated with greater sensitivity [30]. Hence, the lower sensitivity of the NGS method for some HPV genotypes (relative to INNO-LiPA) may be due to the decreased sensitivity of the MY/GP-based primers.
Several HPV detection methods primarily involve genotyping based on nested-PCR amplification using consensus primers located in the L1 gene followed by HPV type identification by oligonucleotide probe hybridization or direct sequencing. Sanger sequencing methods have the potential to identify a broad range of HPV types, although distinguishing multiple infections may be problematic. Hybridization-based methods can discriminate HPV types in multiple infections, but can only identify HPV types represented by the probes [31]. The advantage of the NGS method over hybridization-based methods is high specificity genotyping, because the method is based on massively parallel sequencing and the results are less susceptible to misinterpretation. The other advantage of NGS is that it can detect multiple infections. Multiple infections are common, but have no additive or synergistic effects on the development of high-risk cervical cancer. In fact, reduced high-risk cervical cancer rates have been correlated with multiple infection-, rather than single-genotype infection profiles, suggesting possible intergenotypic competition or more effective immune responses triggered by multiple infections [32].
A single NGS run can generate millions of DNA sequence reads in 24 hours, which is more than can be achieved with hundreds of Sanger type sequencers over the same time period [30]. However, compared with conventional HPV typing methods, NGS has a longer turnaround time per sample and higher costs, mainly associated with the instrument (Table 2). The NGS technique will improve with automation and standardization of protocols. Moreover, NGS costs decrease with increasing data throughput. With the development of less expensive instruments, NGS may soon become a cost-effective platform for molecular diagnosis of HPV.
A limitation of this study was the small study population. Additionally, depending on the method used and the purity of DNA, different internal controls were used. Therefore, the quantity of amplified DNA differed. Moreover, the NGS method used in this study had an intrinsic drawback because the primers used were based on the MY/GP primers, which have low sensitivity for some HPV genotypes [33]. However, our results show that NGS is a promising method for HPV genotyping because of its high sensitivity for multiple infections and its ability to detect a wide range of HPV genotypes.
Detection of HPV DNA is a useful tool in the early diagnosis of cervical pre-cancer when used in conjunction with cytology. NGS is an alternative technique for carcinogenic HPV detection. This technique has high sensitivity for multiple infections and offers the potential to detect a broad spectrum of HPV genotypes.
Acknowledgements
This work was supported by the Research Chair Grant from NSTDA [p-15-50004]; Dutsadi Piphat Scholarship; the Center of Excellence in Clinical Virology, Chulalongkorn University [GCE 59-00930-005).
References
1. Arbyn M, Castellsagué X, de Sanjosé S, Bruni L, Saraiya M, Bray F, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011; 22:2675–2686. PMID: 21471563.
2. Allan B, Marais DJ, Hoffman M, Shapiro S, Williamson AL. Cervical human papillomavirus (HPV) infection in South African women: implications for HPV screening and vaccine strategies. J Clin Microbiol. 2008; 46:740–742. PMID: 17977997.
3. Bosch FX, Burchell AN, Schiffman M, Giuliano AR, de Sanjose S, Bruni L, et al. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine. 2008; 26:K1–K16. PMID: 18847553.
4. de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004; 324:17–27. PMID: 15183049.
5. Chen L, Watanabe K, Haruyama T, Kobayashi N. Simple and rapid human papillomavirus genotyping method by restriction fragment length polymorphism analysis with two restriction enzymes. J Med Virol. 2013; 85:1229–1234. PMID: 23918541.
6. de Antonio JC, Fernández-Olmos A, Mercadillo M, Lindemann ML, Mochales FB. Detection of high-risk human papillomavirus by two molecular techniques: hybrid capture and linear array. J Virol Methods. 2008; 149:163–166. PMID: 18328575.
7. Manos MM, Ting Y, Wright DK, Lewis AJ, Broker TR, Wolinsky SM. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989; 7:209–214.
8. de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3' ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995; 76:1057–1062. PMID: 9049358.
9. Fuessel Haws AL, He Q, Rady PL, Zhang L, Grady J, Hughes TK, et al. Nested PCR with the PGMY09/11 and GP5(+)/6(+) primer sets improves detection of HPV DNA in cervical samples. J Virol Methods. 2004; 122:87–93. PMID: 15488625.
10. van Hamont D, van Ham MA, Bakkers JM, Massuger LF, Melchers WJ. Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the roche linear array HPV genotyping test. J Clin Microbiol. 2006; 44:3122–3129. PMID: 16954236.
11. Coutlée F, Rouleau D, Petignat P, Ghattas G, Kornegay JR, Schlag P, et al. Enhanced detection and typing of human papillomavirus (HPV) DNA in anogenital samples with PGMY primers and the Linear array HPV genotyping test. J Clin Microbiol. 2006; 44:1998–2006. PMID: 16757590.
12. Chansaenroj J, Theamboonlers A, Chinchai T, Junyangdikul P, Swangvaree S, Karalak A, et al. High-risk human papillomavirus genotype detection by electrochemical DNA chip method. Asian Pac J Cancer Prev. 2012; 13:1151–1158. PMID: 22799297.
13. Radford AD, Chapman D, Dixon L, Chantrey J, Darby AC, Hall N. Application of next-generation sequencing technologies in virology. J Gen Virol. 2012; 93:1853–1868. PMID: 22647373.
14. Sun C, McAndrew T, Smith BC, Chen Z, Frimer M, Burk RD. Characterization of HPV DNA methylation of contiguous CpG sites by bisulfite treatment and massively parallel sequencing-the FRAGMENT approach. Front Genet. 2014; 5:150. PMID: 24917876.
15. Yi X, Zou J, Xu J, Liu T, Liu T, Hua S, et al. Development and validation of a new HPV genotyping assay based on next-generation sequencing. Am J Clin Pathol. 2014; 141:796–804. PMID: 24838323.
16. Chinchai T, Chansaenroj J, Junyangdikul P, Swangvaree S, Karalak A, Niruthisard S, et al. Comparison between direct sequencing and INNO-LiPA methods for HPV detection and genotyping in Thai Women. Asian Pac J Cancer Prev. 2011; 12:989–994. PMID: 21790239.
17. Lurchachaiwong W, Junyangdikul P, Payungporn S, Chansaenroj J, Sampatanukul P, Tresukosol D, et al. Relationship between hybrid capture II ratios and DNA amplification of E1, E6 and L1 genes used for the detection of human papillomavirus in samples with different cytological findings. Asian Pac J Allergy Immunol. 2009; 27:217–224. PMID: 20232576.
18. Ermel A, Qadadri B, Morishita A, Miyagawa I, Yamazaki G, Weaver B, et al. Human papillomavirus detection and typing in thin prep cervical cytologic specimens comparing the Digene Hybrid Capture II Assay, the Roche Linear Array HPV Genotyping Assay, and the Kurabo GeneSquare Microarray Assay. J Virol Methods. 2010; 169:154–161. PMID: 20670658.
19. Barzon L, Militello V, Lavezzo E, Franchin E, Peta E, Squarzon L, et al. Human papillomavirus genotyping by 454 next generation sequencing technology. J Clin Virol. 2011; 52:93–97. PMID: 21802982.
20. Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008; 5:16–18. PMID: 18165802.
21. Smith BC, McAndrew T, Chen Z, Harari A, Barris DM, Viswanathan S, et al. The cervical microbiome over 7 years and a comparison of methodologies for its characterization. PLoS One. 2012; 7:e40425. PMID: 22792313.
22. Cheng YP, Chen CW, Sheen YS, Tsai TF. Genotype distribution of human papillomavirus in anogenital warts of male patients in Taiwan. Derm Sinica. 2012; 30:85–89.
23. Menzo S, Monachetti A, Trozzi C, Ciavattini A, Carloni G, Varaldo PE, et al. Identification of six putative novel human papillomaviruses (HPV) and characterization of candidate HPV type 87. J Virol. 2001; 75:11913–11919. PMID: 11689676.
24. Maver PJ, Kocjan BJ, Seme K, Poljak M. Genomic diversity of low-risk human papillomavirus genotypes HPV 40, HPV 42, HPV 43, and HPV 44. J Med Virol. 2014; 86:272–282. PMID: 24155245.
25. Sasagawa T, Basha W, Yamazaki H, Inoue M. High-risk and multiple human papillomavirus infections associated with cervical abnormalities in Japanese women. Cancer Epidemiol Biomarkers Prev. 2001; 10:45–52. PMID: 11205488.
26. International Agency for Research on Cancer. IARC monographs on the evaluation of carcinogenic risks to humans. Human papillomaviruses, volume 90. Lyon, France: International Agency for Research on Cancer;2007.
27. Muñoz N, Castellsagué X, de González AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006; 24:S3/1-10.
28. Micalessi MI, Boulet GA, Bogers J. A real-time PCR approach based on SPF10 primers and the INNO-LiPA HPV genotyping extra assay for the detection and typing of human papillomavirus. Methods Mol Biol. 2015; 1249:27–35. PMID: 25348295.
29. Perrons C, Kleter B, Jelley R, Jalal H, Quint W, Tedder R. Detection and genotyping of human papillomavirus DNA by SPF10 and MY09/11 primers in cervical cells taken from women attending a colposcopy clinic. J Med Virol. 2002; 67:246–252. PMID: 11992586.
30. da Fonseca AJ, Galvão RS, Miranda AE, Ferreira LC, Chen Z. Comparison of three human papillomavirus DNA detection methods: next generation sequencing, multiplex-PCR and nested-PCR followed by Sanger based sequencing. J Med Virol. 2016; 88:888–894. PMID: 26496186.
31. Abreu AL, Souza RP, Gimenes F, Consolaro ME. A review of methods for detect human Papillomavirus infection. Virol J. 2012; 9:262. PMID: 23131123.
32. Salazar KL, Zhou HS, Xu J, Peterson LE, Schwartz MR, Mody DR, et al. Multiple human papilloma virus infections and their impact on the development of high-risk cervical lesions. Acta Cytol. 2015; 59:391–398. PMID: 26674365.
33. Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlée F, Hildesheim A, et al. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000; 38:357–361. PMID: 10618116.
Table 1
Summary of next-generation sequencing data generated from each run
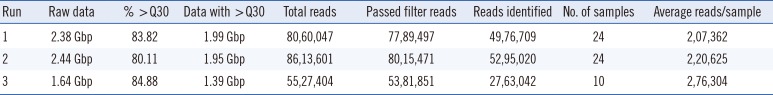
Table 2
Summary of four HPV genotyping methods (HPV major type and minor types detected by next-generation sequencing [NGS] for all samples/percentage of total reads obtained in the sequencing)




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 >30%
>30%  20–29.9%
20–29.9%  10–19.9%
10–19.9%  1–9.9%
1–9.9%  <1%
<1% XML Download
XML Download