Abstract
Background
The interaction between killer immunoglobulin-like receptors (KIRs) and HLA class I regulates natural killer (NK) cell cytotoxicity and function. The impact of NK cell alloreactivity through KIR in liver transplantation remains unelucidated. Since the frequency of HLA-C and KIR genotypes show ethnic differences, we assessed the impact of HLA-C, KIR genotype, or KIR-ligand mismatch on the allograft outcome of Korean liver allografts.
Methods
One hundred eighty-two living donor liver transplant patients were studied. Thirty-five patients (19.2%) had biopsy-confirmed acute rejection (AR), and eighteen (9.9%) had graft failure. The HLA-C compatibility, KIR genotypes, ligand-ligand, and KIR-ligand matching was retrospectively investigated for association with allograft outcomes.
Results
Homozygous C1 ligands were predominant in both patients and donors, and frequency of the HLA-C2 allele in Koreans was lower than that in other ethnic groups. Despite the significantly lower frequency of the HLA-C2 genotype in Koreans, donors with at least one HLA-C2 allele showed higher rates of AR than donors with no HLA-C2 alleles (29.2% vs 15.7%, P=0.0423). Although KIR genotypes also showed ethnic differences, KIR genotypes and the number of activating KIR/inhibitory KIR were not associated with the allograft outcome. KIR-ligand mismatch was expected in 31.6% of Korean liver transplants and had no impact on AR or graft survival.
Natural killer (NK) cells may participate in solid-organ transplantation rejection by modulating host immune reaction rather than direct effector function [123]. NK cell effector function is dependent on the interaction between killer cell immunoglobulin-like receptors (KIRs) and their ligands, HLA class I molecules. NK cells sense the loss of self cell-surface HLA class I molecules and trigger cytolysis to target infected or transformed cells [4]. NK cells express a variety of receptors, including the KIR, and KIRs may be either activating (aKIRs) or inhibitory (iKIRs) [56]. HLA-C is a major ligand for KIR, and HLA-C molecules can be divided into two groups—HLA-C1 ligand or HLA-C2 ligand—based on their KIR specificity. The HLA-C1 ligand carries an asparagine at position 80 (HLA-Cw01, 03, 07, 08, 09, 10, 12, 14) and HLA-C2 has a lysine at position 80 (HLA-Cw02, 04, 05, 06, 15) [7]. Within iKIRs, KIR2DL1 recognizes HLA-C2 ligand, whereas KIR2DL2/3 recognizes HLA-C1 ligand [8]. In aKIRs, binding to HLA-C has only been demonstrated for KIR2DS1 and KIR2DS4. KIR2DS1 recognizes HLA-C2 ligand, whereas KIR2DS4 recognizes HLA-C2 and a limited number of HLA-C1 ligands; ligands for other aKIRs remain elusive [8].
The impact of NK cell alloreactivity through KIR in solid organ transplantation remains unclear. Previous studies utilized several KIR models such as KIR gene-gene mismatch, HLA-C genotype mismatch, and KIR-ligand mismatch for donors lacking ligands for inhibitory KIR, which present in recipients [589]. In liver transplantation (LT), previous studies reported the impact of HLA-C allotypes on acute rejection (AR) and graft survival [910111213], but some studies demonstrated no effect on graft outcomes [14]. In addition, most previous studies for HLA-C, KIR genotypes, or KIR-ligand matching in LT do not report Asians as a separate group, and no data has been acquired previously for Korean liver transplants. Since the frequency of HLA-C genotypes and KIR genotypes differs with ethnicity [15], we assessed the impact of HLA-C genotypes, KIR genotype, or KIR-ligand mismatch on the clinical outcome of Korean liver allografts.
This study included 182 patients who received living donor LT between July 2009 and June 2013 at Seoul St. Mary's Hospital, Korea. AR was defined as clinical suspicion with histopathological evidence or biochemical response to corticosteroid treatment. Biopsy specimens were evaluated by an experienced pathologist and scored according to the Banff scheme [16]. The histopathological evidence of AR was defined as a rejection activity index ≥3. The standard immunosuppressive regimen at our center has previously been described [17]. Demographic characteristics of patients are listed in Table 1. This retrospective study was approved by the Institutional Review Board of Seoul St. Mary's Hospital. The informed consent forms for genetic tests were obtained from the patients.
In all recipients and donors, HLA-A, B, C, and DR typing was performed by DNA molecular typing using sequence-specific oligonucleotide probes (SSOP) with LIFECODES HLA SSO typing kits (Immucor, Stamford, CT, USA). HLA-C alleles were assigned to either C1 or C2 groups, as HLA-C1 carries an asparagine at position 80 (HLA-Cw01, 03, 07, 08, 09, 10, 12, 14) and HLA-C2 has a lysine at position 80 (HLA-Cw02, 04, 05, 06, 15) [7]. HLA-C genotypes in donors and patients grouped into C1C1, C1C2, or C2C2.
KIR genotyping was available for 98 recipients. Genotyping of 16 KIR genes (2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 2DP1, 3DL1, 3DL2, 3DL3, 3DS1, and 3DP1) was performed by the KIR SSO genotyping test (One Lambda, Canoga Park, CA, USA), according to the manufacturer's instructions. The KIR SSO genotyping test applies Luminex technology to the reverse SSOP DNA typing. Three sets of specific primers for exons 3+4, 5, and 7-9 were used for DNA amplification. Each PCR product was biotinylated, which allows later detection using R-phycoerythrin-conjugated streptavidin. PCR products were denatured and hybridized to complementary DNA probes conjugated to fluorescently coded microspheres. After washing the beads, bound amplified DNA was tagged with R-Phycoerythrin-conjugated Streptavidin (SAPE), and a LABScan 100 (One Lambda) was used to measure the fluorescent intensity of each microsphere. Genotype classification was based on the patient's reaction pattern compared to patterns associated with published KIR gene sequences.
aKIRs were typed for 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, and 3DS1, whereas iKIRs were typed for 2DL1, 2DL2, 2DL3, 2DL5, 3DL1, and 3DL2 [18]. One of them, 2DL4, was excluded from the analysis because a previous study reported that it had both activating and inhibitory functions, and 3DL3 was not included because its expression and function remain unclear [5]. Additionally, we grouped the recipient KIR genotypes into AA, BB, and AB according to Tran et al [5]. Patients were classified as AA if none of the following KIRs were present: 2DL2, 2DL5, 3DS1, 2DS1, 2DS2, 2DS3, and 2DS5. For AB and BB genotypes, if the AA genotype was excluded and 2DS4 was absent, the BB genotype was assigned, and the others were designated as AB. To predict NK alloreactivity, we evaluated two main models: the 'ligand-ligand' or 'missing-self' model, and the 'receptor-ligand' or 'missing-ligand' model [581920]. 'Missing-self model' was defined as KIR-ligand mismatching between the donor and recipient, and the effect of HLA-C ligand compatibility on the liver allograft outcome was investigated. The 'missing-ligand' model was defined as the donor lacking ligands for KIR receptors present in the recipient genotype.
Statistical analyses were performed with the statistical package MedCalc version 15.5 (MedCalc, Mariakerke, Belgium). Categorical variables were compared between the two groups by Pearson uncorrected Chi-square test or Fisher's exact test. Graft survival and patient survival rates were analyzed by Kaplan-Meier survival and Cox regression methods. All reported P values are two-sided, and P values <0.05 were considered statistically significant.
Thirty-five patients (19.2%) exhibited biopsy-confirmed AR and 18 (9.9%) exhibited graft failure. The median (95% confidence interval) duration of graft survival and patient survival were 40.0 (36.0-43.0) months and 40.5 (36.0-43.0) months, respectively. There were no significant differences with regard to age, gender, HLA-A/B/DR mismatches, underlying diseases, crossmatching positivity, or immunosuppression between recipients with or without AR of liver allograft. In addition, there were no independent risk factors for graft survival.
In the study population, the homozygous C1 genotype was predominant in both patients and donors (Table 2), and the homozygous C2 genotype was found in only five (2.7%) recipients and two (1.1%) donors. When we investigated the independent effect of donor and recipient HLA-C genotypes, there was no significant difference in AR incidence between HLA-C1C1, HLA-C1C2, and HLA-C2C2 genotypes of recipients or donors. In terms of the HLA-C allele match or mismatch, HLA-C allelic compatibility was not associated with AR development (0 mismatch, 22.7%; 1 mismatch, 21.3%; 2 mismatch, 14.3%). When we investigated the HLA-C ligand disparity between donors and recipients for the missing self model, which implies absence of ligands in the donor for the inhibitory receptors that are present in the recipients, HLA-C ligand disparity had no influence on AR rate (5/26 [19.2%] vs 30/156 [19.2%]). However, the donor HLA-C ligand group had an effect on AR. The incidence of AR increased as the number of donor HLA-C2 alleles increased (C1C1, 15.7%; C1C2 28.3%; C2C2 50.0%) (Chi-square for trend 4.636, P=0.0313). AR was more frequently observed in donors with at least one HLA-C2 allele compared with HLA-C1 homozygous donors (14/48 vs 21/134, P=0.0423) (Fig. 1). However, no significant effect of donor HLA-C2 was observed on graft survival or patient survival (Fig. 2).
The frequencies of KIR genes and KIR gene distribution are shown in Fig. 3. KIR2DL1, KIR2DL3, KIR2DL4, KIR2DP1, KIR3DL2, KIR3DL3, and KIR3DP1 were detected in all patients. KIR2DL2, KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DL1, and KIR3DS1 were detected in 15 (15.3%), 34 (34.7%), 34 (34.7%), 16 (16.3%), 13 (13.3%), 92 (93.9%), 25 (25.5%), 92 (93.9 %), and 33(33.7%) patients, respectively. There was no significant difference in the individual KIR gene frequency between the liver allograft AR and AR-free groups. A similar negative result was obtained when we investigated the number of recipients with activating or inhibitory KIR genes. The number of activating or inhibitory KIR genes in the recipient genotype was not associated with liver allograft AR or graft survival.
The 98 patients were categorized into three groups on the basis of KIR-AA, AB, or BB genotypes. The KIR-AA genotype (58.2%) was the most common, followed by AB (35.7%) and BB (6.1%) genotypes. There was no significant difference among the three KIR genotype groups with regard to the development of AR; incidence was 12.3% (7/57) for the AA genotype, 14.3% (5/35) for the AB genotype, and 16.7% (1/6) for the AB genotype. Cox regression analysis also did not show a significant difference in 6-yr allograft survival among the three KIR genotype groups.
We evaluated KIR-HLA ligand mismatches for association with poor liver allograft outcome. KIR-ligand match/mismatch was defined according to the previous study by van Bergen et al [20]. A mismatch for iKIR was defined as the presence of a KIR and its ligand in the recipients and the absence of the same ligand in the donor. A mismatch for aKIR was defined as the presence of the ligand in the donor and the absence of the ligand in the recipient. Among the 98 patients, the patients who were expected to have at least one KIR-ligand mismatch are shown in Table 3. Of them, only one patient developed AR, so we could not confirm the association between KIR-ligand mismatch and AR.
In allogeneic transplantation, NK cells in recipients may be activated through the lack of HLA ligands for iKIRs and the presence of an HLA ligand for an aKIR [20]. We demonstrated that patients who received liver transplants from donors with the HLA-C2 allele had a higher risk of AR than those who received livers from donors with a homozygous HLA-C1 genotype. This finding confirms the previous data by Lopez-Alvarez et al [11], who showed that AR was more frequently detected in C2 homozygous patients in combination of C1/C2 heterozygous donors than in C1 homozygous patients. Similarly, Legaz et al [8] reported that KIR2DL3 and KIR2DS1 in patients increased AR incidence in the presence of donor C2 ligands. Our results support the previous studies suggesting that the KIR2DL1/S1+ allogeneic T cell subset, which is specific for donor C2 ligands, may be induced, or the binding of aKIR (KIR2DS1) to a donor HLA-C2 ligand can induce alloreactivity [2122]. However, other studies demonstrated controversial data showing no impact of donor HLA-C2 genotype on graft or patient survival [91419]. HLA-C proteins may act as alloantigens, which can induce T cell activation and may also function as ligands for KIRs, modulating NK and T cell cytotoxicity [9]. The differences in study results may be due to differences in the study design, ethnic groups examined, diseases leading to LT, cohort size, or other factors, including donor-specific HLA-C antibodies.
The frequency of the HLA-C2 allele is lower in Koreans than in other ethnic groups [15]. In Caucasians, the HLA-C2 allele is predominant and the HLA-C2 homozygous type was detected in about 15% of donors [23]. In the current study, the HLA-C2 homozygous type was found only in five patients (2.7%) and two donors (1.1%) among the Korean cohort. This finding is in agreement with previous reports that showed a HLA-C2 homozygous type frequency of 2.0-3.8% in Koreans [2425]. Despite the significantly low frequency of the HLA-C2 genotype in Koreans, our data demonstrated that the HLA-C2 allele in donors was a risk factor for liver allograft rejection. Although a previous study reported that the KIR2DL1-C2 interaction provides a stronger inhibitory signal to NK cells than KIR2DL2/3-C1 binding [9], we could not confirm this finding in the current study. Further large cohort studies are needed to clarify the clinical impact of KIR gene-ligand interaction. The frequencies of KIR genes and KIR gene distribution were similar to those previously reported in Korean populations (P>0.05) [24]. When we analyzed ethnic differences in KIR gene frequency, Koreans had a significantly lower frequency of KIR2DL2, KIR2DL5, KIR2DS2, and KIR2DS3 genotypes and they had a higher frequency of KIR2DL3 than white populations [8]. These ethnic variations might affect the clinical impact of KIR-ligand mismatch on LT outcomes in Korean recipients. The ethnic differences between the previous study and our data appeared to be due to the underlying cause of LT and the type of donor. In Korea, the predominant disease necessitating LT is hepatitis B virus (HBV)-related liver disease, but in Western countries, hepatitis C virus (HCV)-related liver disease and autoimmunity account for a considerable proportion along with viral hepatitis [1119]. Although direct influence of the underlying disease on clinical outcome was not sufficient, different underlying diseases may affect patient NK cell function.
Since the KIR haplotype B has several activating KIR genes and recipient KIR genotypes (AA, AB, BB) have been investigated in previous studies [526], we analyzed whether recipient KIR genotypes might have an impact on liver allograft outcome. However, we failed to demonstrate any association between the specific KIR genotype group with liver allograft outcome.
There have been conflicting reports on the impact of KIR genes and KIR-ligand matching in LT [910111213]. KIR-ligand mismatches might induce recipient NK alloreactivity toward the liver allograft, resulting in a negative effect on the graft. However, we could not confirm the significance of KIR-ligand matching in liver allografts upon categorizing liver allografts according to KIR-ligand pairs. This may be due to the potentially protective effect of KIR-ligand mismatch during the early post-transplant period, or alternatively, the coexistence of alloreactive NK and T cells in the allograft may cause subclinical graft damage [8]. Our results support the previous suggestion that recipient NK cell lysis of renal graft-derived donor antigen-presenting cells could lead to inhibition of the direct pathway of antigen pathway [5].
We adopted the definition of KIR-ligand mismatch used by Van Bergen et al [20], based on the missing self model via the inhibitory receptors KIR2DL1, KIR2DL2, KIR2DL3, KIR3DL1, and KIR3DL2 [5]. In addition, the activating receptor KIR2DS1 was also included considering that KIR2DS1 binds to HLA-C2 and allows NK cells to respond to allogeneic C2-positive target cells [20]. However, there have been reports on complex interactions between KIR and HLA-C [27], and our study could not demonstrate a clear result. These cross-reactions make it difficult to divide patients into KIR-ligand match vs mismatch groups, and KIR genotyping at the allele level may be required [520].
Overall, based on our results, we identified a meaningful association between HLA-C genotype and AR of liver allograft. The role of NK cells and the biological mechanism of NK-cell alloreactivity in LT are becoming increasingly apparent, and a more cautious review is needed before the clinical application of KIR genotype and KIR-ligand matching for liver allografts. Our study in Korean LT recipients will help to design further studies of NK cell function and assess the impact of KIR ligands on transplantation biology.
Acknowledgments
This research was supported by a grant from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea (A092258).
References
1. Kitchens WH, Uehara S, Chase CM, Colvin RB, Russell PS, Madsen JC. The changing role of natural killer cells in solid organ rejection and tolerance. Transplantation. 2006; 81:811–817. PMID: 16570001.
2. Beilke JN, Kuhl NR, Van Kaer L, Gill RG. NK cells promote islet allograft tolerance via a perforin-dependent mechanism. Nat Med. 2005; 11:1059–1065. PMID: 16155578.
3. Yu G, Xu X, Vu MD, Kilpatrick ED, Li XC. NK cells promote transplant tolerance by killing donor antigen-presenting cells. J Exp Med. 2006; 203:1851–1858. PMID: 16864660.
4. Ljunggren HG. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990; 11:237–244. PMID: 2201309.
5. Tran TH, Unterrainer C, Fiedler G, Döhler B, Scherer S, Ruhenstroth A, et al. No impact of KIR-ligand mismatch on allograft outcome in HLA-compatible kidney transplantation. Am J Transplant. 2013; 13:1063–1068. PMID: 23398855.
6. Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, et al. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. 1995; 182:875–884. PMID: 7650491.
7. Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by "group 2" or "group 1" NK clones. J Exp Med. 1995; 182:605–609. PMID: 7629517.
8. Legaz I, López-Álvarez MR, Campillo JA, Moya-Quiles MR, Bolarín JM, de la, et al. KIR gene mismatching and KIR/C ligands in liver transplantation: consequences for short-term liver allograft injury. Transplantation. 2013; 95:1037–1044. PMID: 23478359.
9. Hanvesakul R, Spencer N, Cook M, Gunson B, Hathaway M, Brown R, et al. Donor HLA-C genotype has a profound impact on the clinical outcome following liver transplantation. Am J Transplant. 2008; 8:1931–1941. PMID: 18671674.
10. Moya-Quiles MR, Alvarez R, Miras M, Gomez-Mateo J, Lopez-Alvarez MR, Marin-Moreno I, et al. Impact of recipient HLA-C in liver transplant: a protective effect of HLA-Cw*07 on acute rejection. Hum Immunol. 2007; 68:51–58. PMID: 17207712.
11. López-Alvarez MR, Moya-Quiles MR, Minguela A, Gil J, Miras M, Campillo JA, et al. HLA-C matching and liver transplants: donor-recipient genotypes influence early outcome and CD8+KIR2D+ T-cells recuperation. Transplantation. 2009; 88(3 Suppl):S54–S61. PMID: 19667963.
12. Moya-Quiles MR, Muro M, Torío A, Sánchez-Bueno F, Miras M, Marín L, et al. Human leukocyte antigen-C in short- and long-term liver graft acceptance. Liver Transpl. 2003; 9:218–227. PMID: 12619017.
13. Bishara A, Brautbar C, Zamir G, Eid A, Safadi R. Impact of HLA-C and Bw epitopes disparity on liver transplantation outcome. Hum Immunol. 2005; 66:1099–1105. PMID: 16571410.
14. Tran TH, Middleton D, Döhler B, Scherer S, Meenagh A, Sleator C, et al. Reassessing the impact of donor HLA-C genotype on long-term liver transplant survival. Am J Transplant. 2009; 9:1674–1678. PMID: 19392983.
15. Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernández-Viña MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2001; 62:1009–1030. PMID: 11543903.
16. Banff schema for grading liver allograft rejection: an international consensus
document. Hepatology. 1997; 25:658–663. PMID: 9049215.
17. Na GH, Kim EY, Hong TH, You YK, Kim DG. Effects of preoperative positive cross-match and HLA mismatching on early acute cellular rejection and graft survival in living donor liver transplantation. Ann Transplant. 2015; 20:553–560. PMID: 26377312.
18. Gagne K, Brizard G, Gueglio B, Milpied N, Herry P, Bonneville F, et al. Relevance of KIR gene polymorphisms in bone marrow transplantation outcome. Hum Immunol. 2002; 63:271–280. PMID: 12039408.
19. Moroso V, van der Meer A, Tilanus HW, Kazemier G, van der Laan LJ, Metselaar HJ, et al. Donor and recipient HLA/KIR genotypes do not predict liver transplantation outcome. Transpl Int. 2011; 24:932–942. PMID: 21672051.
20. van Bergen J, Thompson A, Haasnoot GW, Roodnat JI, de Fijter JW, Claas FH, et al. KIR-ligand mismatches are associated with reduced long-term graft survival in HLA-compatible kidney transplantation. Am J Transplant. 2011; 11:1959–1964. PMID: 21714849.
21. Chewning JH, Gudme CN, Hsu KC, Selvakumar A, Dupont B. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J Immunol. 2007; 179:854–868. PMID: 17617576.
22. Foley B, De Santis D, Lathbury L, Christiansen F, Witt C. KIR2DS1-mediated activation overrides NKG2A-mediated inhibition in HLA-C C2-negative individuals. Int Immunol. 2008; 20:555–563. PMID: 18308713.
23. Michelo CM, van der Meer A, Tijssen HJ, Zomer R, Stelma F, Hilbrands LB, et al. KIR and Human Leukocyte Antigen genotype associated risk of cytomegalovirus disease in renal transplant patients. Transplantation. 2015; 99:1506–1513. PMID: 25427165.
24. Kim HJ, Choi HB, Jang JP, Baek IC, Choi EJ, Park M, et al. HLA-Cw polypmorphism and killer cell immunoglobulin-like receptor (KIR) gene analysis in Korean colorectal cancer patients. Int J Surg. 2014; 12:815–820. PMID: 24998207.
25. Park H, Rho EY, In JW, Kim I, Yoon SS, Park S, et al. The impact of HLA and KIR ligand mismatching on unrelated allogeneic hematopoietic stem cell transplantation in Korean adult patients. Ann Lab Med. 2015; 35:111–117. PMID: 25553290.
26. Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2009; 113:726–732. PMID: 18945962.
27. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008; 180:3969–3979. PMID: 18322206.
Fig. 1
Data represent the incidence of acute rejection (AR) in living donor liver transplant patients according to the donor HLA-C groups. (A) The incidence of AR increased as the number of HLA-C2 alleles increased (chi-square for trend 4.636, P=0.0313). (B) The AR incidence in patients with and without a donor HLA-C2 allele showed a statistically significant difference (P=0.0423).
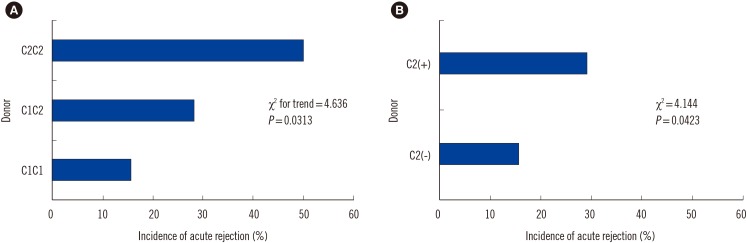
Fig. 2
Comparison of graft survival (A) and patient survival (B) between HLA-C2 allele negative and HLA-C2 allele positive donors showed no significant difference.
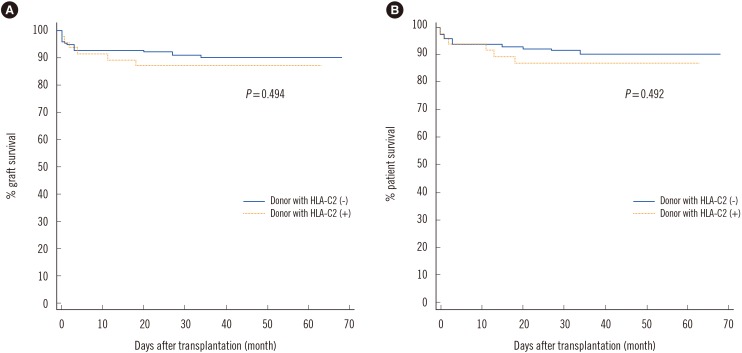
Fig. 3
(A) KIR gene distribution with or without acute rejection. (B) KIR genotype patterns in patients with acute rejection and the total population.
Abbreviation: KIR, killer immunoglobulin-like receptor.
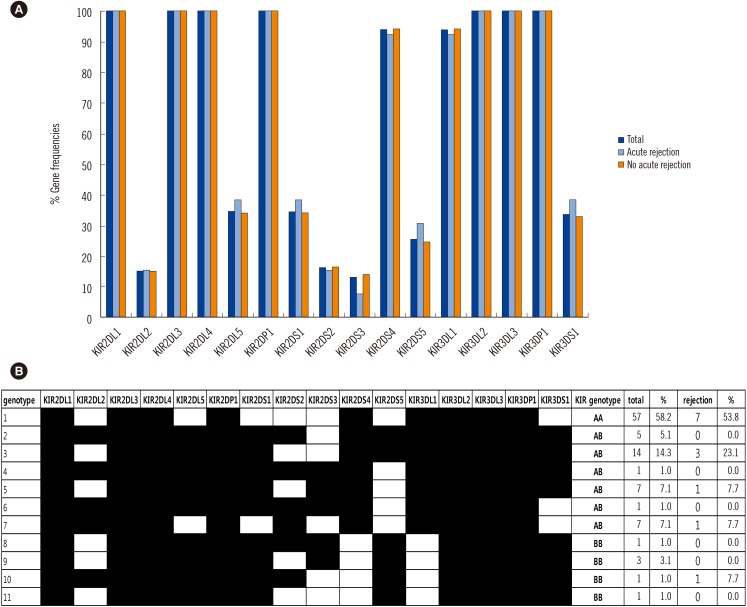
Table 1
Demographic data of the study population
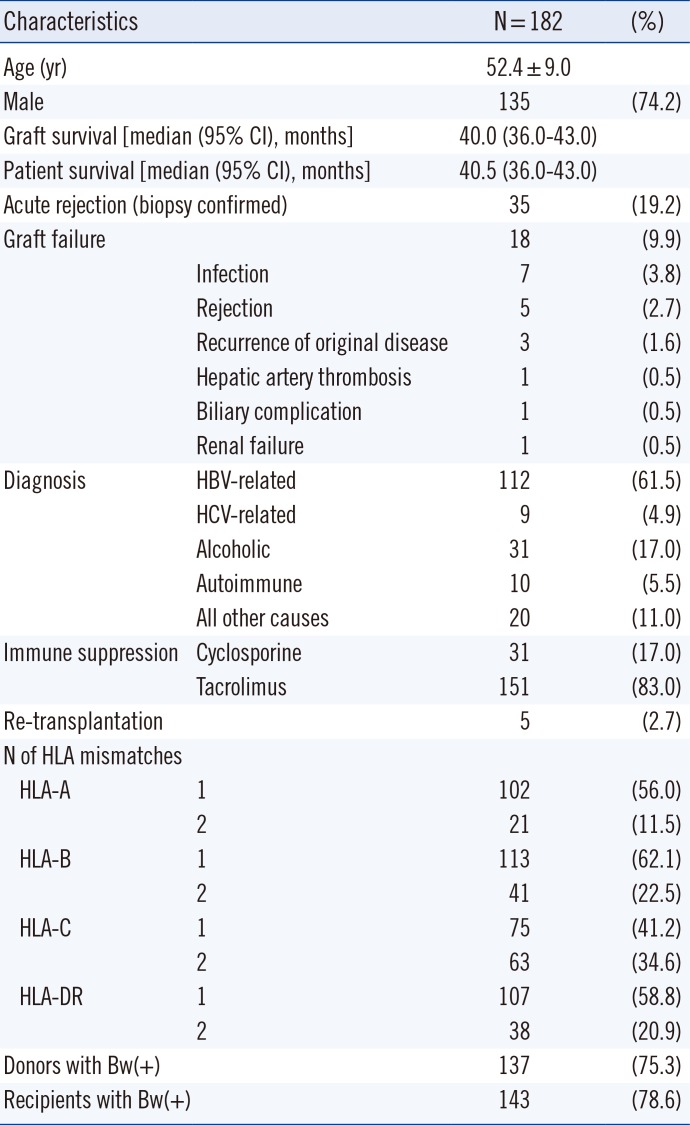
Table 2
HLA-C genotype combinations in the study population
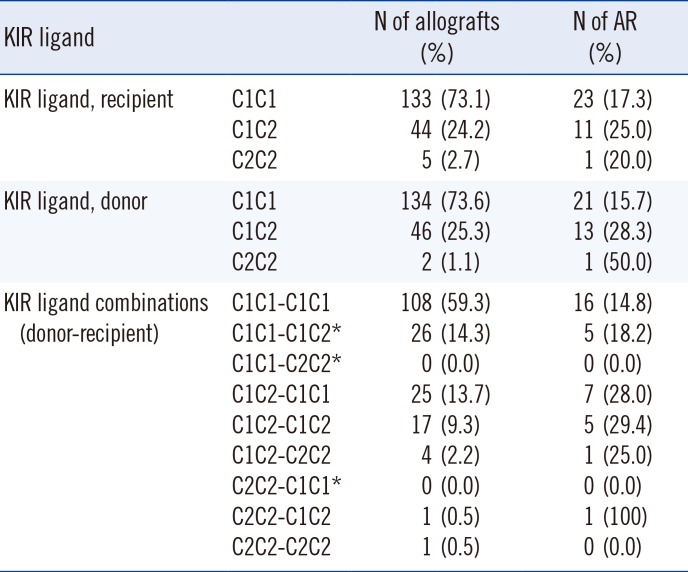
Table 3
KIR-ligand match/mismatch
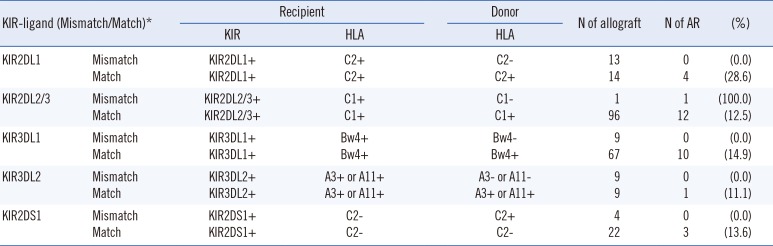
*KIR-ligand match/mismatch was defined according to van Bergan et al [20].
Abbreviations: KIR, killer immunoglobulin-like receptor; AR, acute rejection.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download