Abstract
Hereditary gelsolin amyloidosis (HGA) is an autosomal dominant hereditary disease characterized by corneal lattice dystrophy, peripheral neuropathy, and cutis laxa. So far, no Korean patients with HGA have been reported. A 58-yr-old man presented with involuntary facial twitching, progressive bilateral facial weakness, and tongue atrophy. His mother, maternal uncle, two sisters, and son suffered from the same symptoms. Electrophysiological studies revealed signs of chronic denervation in the cervical and lumbar regions, mild sympathetic autonomic dysfunction, and bilateral facial nerve dysfunction. Diagnostic whole-exome sequencing (WES) revealed a p.D214Y heterozygous mutation in the gelsolin gene in affected members. We present the first report of a Korean family with HGA diagnosed by WES. WES facilitated a clinical diagnosis of HGA in patients with undiagnosed neuropathies.
Hereditary gelsolin amyloidosis (HGA; also known as familial amyloidosis of the Finnish type, or FAF) is an autosomal dominant amyloidosis [1]. Clinical manifestations of HGA begin in the third or fourth decade of life with corneal lattice dystrophy, followed by cutis laxa and slowly progressive neuropathy [1234567]. Since the first report from Finland, additional cases of HGA have been identified in many ethnic groups [13456891011121314]. However, no Korean patients with HGA have been reported.
So far, four mutations in the gelsolin gene (GSN) have been identified as causes of HGA. The p.D214N mutation, the first discovered and the most common type, was identified in patients with typical presentations and complete penetrance [13456891013]. The second mutation discovered, p.D214Y, also leads to HGA that is clinically similar to that caused by p.D214N [56111214]. The p.D214N mutation was identified in Finnish, Japanese, American, Dutch, Portuguese, British, and Iranian families, and the p.D214Y mutation was found in Danish, Czech, and Brazilian families [13456891011121314]. It has been suggested that the p.D214Y mutation occurs irrespective of ethnicity, and that there are multiple founders [4713]. Recently, two additional mutations, p.G194R and p.N211K, were reported to be associated with renal amyloidosis in patients without corneal lattice dystrophy or neuropathies [121516]. We present the first report of a Korean family with HGA caused by p.D214Y mutation.
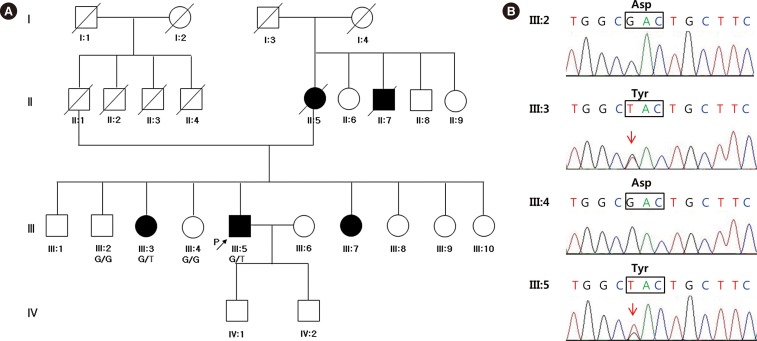
A 58-yr-old Korean male presented with involuntary facial twitching and weakness, loose skin, and a deep-furrowed tongue. The symptoms had been slowly progressing since he was 54 yr old. The patient's mother, maternal uncle, and two sisters suffered from the same symptoms (Fig. 1A). This Korean family has no non-Korean relatives. The proband complained of paresthesia of the right hand, intermittent cramps on the right calf, and decreased sweating on both hands. He had no ophthalmic complaints. Neurological examination revealed mild weakness in the orbicularis oculi and orbicularis oris muscles; however, no clear motor weakness or sensory changes were observed in the four extremities.
We performed electrophysiologic studies, including nerve conduction study (NCS), electromyography (EMG), and autonomic function tests (AFTs). In the facial NCS and EMG, reduced compound muscle action potential (CMAP) amplitudes and conduction velocity of the left facial nerve and reduced CMAP amplitude of the right facial nerve were identified. In addition, blink reflex tests showed borderline to mildly prolonged latencies of ipsilateral R1 and R2 and contralateral R2 waves with facial nerve stimulations on either side. Routine NCSs on the right extremities revealed reduced CMAP amplitude and conduction velocity and delayed terminal latency in the right peroneal nerve, and a mildly reduced distal sensory nerve action potential (SNAP) in the right median nerve. EMG of the right first dorsal interosseous muscle and tibialis anterior muscle revealed reduced recruitment of motor unit action potential. In AFTs, sympathetic skin responses to auditory, tactile, and electrical stimuli were missing in hands and feet, without evident abnormalities in heart rate variability, Valsalva ratio, Valsalva maneuver, or cardiovascular response to standing. Taken together, the electrophysiological studies demonstrated signs of bilateral facial nerve dysfunction, chronic denervation of the cervical and lumbar regions, and sudomotor dysfunction.
A trinucleotide repeat expansion analysis yielded no remarkable findings, excluding Kennedy disease. To identify the etiology of the undiagnosed neuropathy, we performed whole-exome sequencing (WES) following written informed consent.
Germline DNA was obtained from the four individuals (Fig. 1A). The exomes were targeted by using an SureSelect 50 Mb All Exon Kit (Agilent, Santa Clara, CA, USA) and sequenced on an HiSeq 2000 (Illumina, San Diego, CA, USA) (2×100 bp) according to the manufacturer's instructions. We generated an average of 78,148,365 reads resulting in an average of 6,989,878,321 high quality (≥MQ20) bases per sample. The average depth of the on-target regions was 90 fold. The reads were mapped to hg19 by using the Burrow-Wheeler Aligner (BWA 0.6.2.-r126, Wellcome Trust Sanger Institute, Cambridge, UK), and duplication removal and recalibrations were performed by using Picard Tools 1.64 (Wellcome Trust Sanger Institute) and GATK-Lite (Broad Institute, Inc, Cambridge, MA, USA), respectively. A total of 55,199 variants were called and functionally annotated by using ANNOVAR [1718]. To prioritize the variants, we applied the following criteria: allele frequency <0.01 in the 1000 Genome Project (http://browser.1000genomes.org/index.html) and the Exome Sequencing Project (http://evs.gs.washington.edu/EVS/), and mutations predicted to have "deleterious" effects based on SIFT, Polyphen-2 LRT, Mutation-Taster, MutationAssessor, FATHMM, and GERP [18]. A total of 20 variants were present in the affected individuals but not in unaffected individuals. Among them, the p.D214Y mutation in GSN, a previously reported HGA mutation, was selected and validated by Sanger sequencing (Fig. 1B).
We present the first Korean family with HGA diagnosed by a family study and WES. Although the phenotypic manifestations of HGA were known to be typical, some variations have been reported [412151619]. A traditional, phenotype-first approach based on detailed clinical examination is inexpensive and readily available in the clinic. In clinically suspected cases, HGA can be diagnosed by verifying the presence of gelsolin amyloid in the endoneurium or skin. A phenotype-first approach, however, might allow HGA to go undetected in patients. Recently, WES has been successfully applied as a diagnostic tool in the identification of causative mutations in undiagnosed patients [20]. In spite of the diagnostic utility of WES, there are some drawbacks; for example, interpretation of the results can be challenging. To address this, a variant prioritization strategy based on a comprehensive annotation and segregation analysis can help to pinpoint causal variants. While a biopsy is invasive and may require a strong clinical indication, WES is less invasive and is a useful tool in a genotype-first approach.
After identification of the causative mutation, we were unable to confirm the phenotype, corneal lattice dystrophy, associated with this genotype, because the patient was lost to follow-up. However, other clinical manifestations and the results of the electrophysiologic studies were compatible with HGA. Despite the fact that no ophthalmic issues were presented and no eye exam was performed, WES successfully facilitated the clinical diagnosis of HGA. This is the first report of a Korean family with HGA, with cases genetically confirmed by WES.
References
1. Meretoja J. Familial systemic paramyloidosis with lattice dystrophy of the cornea, progressive cranial neuropathy, skin changes and various internal symptoms. A previously unrecognized heritable syndrome. Ann Clin Res. 1969; 1:314–324. PMID: 4313418.
2. Meretoja J. Genetic aspects of familial amyloidosis with corneal lattice dystrophy and cranial neuropathy. Clin Genet. 1973; 4:173–185. PMID: 4543600.

3. Maury CP, Kere J, Tolvanen R, de la Chapelle A. Finnish hereditary amyloidosis is caused by a single nucleotide substitution in the gelsolin gene. FEBS Lett. 1990; 276:75–77. PMID: 2176164.

4. Taira M, Ishiura H, Mitsui J, Takahashi Y, Hayashi T, Shimizu J, et al. Clinical features and haplotype analysis of newly identified Japanese patients with gelsolin-related familial amyloidosis of Finnish type. Neurogenetics. 2012; 13:237–243. PMID: 22622774.

5. Maury CP, Liljeström M, Boysen G, Törnroth T, de la Chapelle A, Nurmiaho-Lassila EL. Danish type gelsolin related amyloidosis: 654G-T mutation is associated with a disease pathogenetically and clinically similar to that caused by the 654G-A mutation (familial amyloidosis of the Finnish type). J Clin Pathol. 2000; 53:95–99. PMID: 10767822.

6. de la Chapelle A, Tolvanen R, Boysen G, Santavy J, Bleeker-Wagemakers L, Maury CP, et al. Gelsolin-derived familial amyloidosis caused by asparagine or tyrosine substitution for aspartic acid at residue 187. Nat Genet. 1992; 2:157–160. PMID: 1338910.

7. Solomon JP, Page LJ, Balch WE, Kelly JW. Gelsolin amyloidosis: genetics, biochemistry, pathology and possible strategies for therapeutic intervention. Crit Rev Biochem Mol Biol. 2012; 47:282–296. PMID: 22360545.

8. Ikeda M, Mizushima K, Fujita Y, Watanabe M, Sasaki A, Makioka K, et al. Familial amyloid polyneuropathy (Finnish type) in a Japanese family: Clinical features and immunocytochemical studies. J Neurol Sci. 2007; 252:4–8. PMID: 17097682.

9. Lüttmann RJ, Teismann I, Husstedt IW, Ringelstein EB, Kuhlenbäumer G. Hereditary amyloidosis of the Finnish type in a German family: clinical and electrophysiological presentation. Muscle Nerve. 2010; 41:679–684. PMID: 20229579.

10. Ardalan MR, Shoja MM, Kiuru-Enari S. Amyloidosis-related nephrotic syndrome due to a G654A gelsolin mutation: the first report from the Middle East. Nephrol Dial Transplant. 2007; 22:272–275. PMID: 16998221.

11. de la Chapelle A, Kere J, Sack GH Jr, Tolvanen R, Maury CP. Familial amyloidosis, Finnish type: G654----a mutation of the gelsolin gene in Finnish families and an unrelated American family. Genomics. 1992; 13:898–901. PMID: 1322359.
12. Chastan N, Baert-Desurmont S, Saugier-Veber P, Dérumeaux G, Cabot A, Frébourg T, et al. Cardiac conduction alterations in a French family with amyloidosis of the Finnish type with the p.Asp187Tyr mutation in the GSN gene. Muscle Nerve. 2006; 33:113–119. PMID: 16258946.
13. Paunio T, Sunada Y, Kiuru S, Makishita H, Ikeda S, Weissenbach J, et al. Haplotype analysis in gelsolin-related amyloidosis reveals independent origin of identical mutation (G654A) of gelsolin in Finland and Japan. Hum Mutat. 1995; 6:60–65. PMID: 7550233.
14. Solari HP, Ventura MP, Antecka E, Belfort Junior R, Burnier MN Jr. Danish type gelsolin-related amyloidosis in a Brazilian family: case reports. Arq Bras Oftalmol. 2011; 74:286–288. PMID: 22068858.

15. Sethi S, Theis JD, Quint P, Maierhofer W, Kurtin PJ, Dogan A, et al. Renal amyloidosis associated with a novel sequence variant of gelsolin. Am J Kidney Dis. 2013; 61:161–166. PMID: 22938848.

16. Efebera YA, Sturm A, Baack EC, Hofmeister CC, Satoskar A, Nadasdy T, et al. Novel gelsolin variant as the cause of nephrotic syndrome and renal amyloidosis in a large kindred. Amyloid. 2014; 21:110–112. PMID: 24601799.

17. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164. PMID: 20601685.

18. Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013; 34:E2393–E2402. PMID: 23843252.

19. Tanskanen M, Paetau A, Salonen O, Salmi T, Lamminen A, Lindsberg P, et al. Severe ataxia with neuropathy in hereditary gelsolin amyloidosis: a case report. Amyloid. 2007; 14:89–95. PMID: 17453628.

20. Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013; 369:1502–1511. PMID: 24088041.

Fig. 1
A Korean family with hereditary gelsolin amyloidosis. (A) Pedigree of the family. The proband is represented by the arrow. Affected individuals are indicated by solid symbols. Germline DNA was obtained from four individuals (III:2, III:3, III:4, and III:5), including the proband. G, reference allele; T, mutant allele. (B) Validation of the mutation by Sanger sequencing. The p.D214Y mutation was identified in a symptomatic sister (III:3) and in the proband (III:5). Sequences identical to the reference genotype were observed in two asymptomatic members (III:2 and III:4) of the family.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download