Abstract
Intermediate-resolution HLA-DQ typing has gained importance in organ transplantation recently. We evaluated the performance of the LIFECODES HLA-DQB1 typing kit (Immucor, USA) using sequence-specific oligonucleotide (SSO) probe and Luminex platform (Luminex Corp., USA) on 100 samples tested by sequence-based typing (SBT) using the AlleleSEQR HLA-DQB1 kit (Abbott Molecular, USA) in Korean individuals. No sample showed ambiguity in the assignment of 4-digit HLA-DQB1 allele with the LIFECODES HLA-DQB1 SSO typing kit, and the results were fully concordant with those of high-resolution typing of AlleleSEQR HLA-DQB1 SBT up to 4-digit level. Three samples required adjustment of false reactions (3/100, 3.0%): two samples with DQB1*03:03/*06:01 showed false-positive result in probe 253, and 1 sample with DQB1*04:02/*05:02 showed false-negative result in probe 217. We tested an additional sample with DQB1*03:03/*06:01, which showed same false-positivity in probe 253 and 2 samples with DQB1*04:02/*05:02, which showed no false reaction. The false reactions did not result in ambiguity or change in the HLA allele assignment. We could assign HLA-DQB1 alleles to 4 digit-level without ambiguity, with 100% concordance with the SBT results. Thus, LIFECODES HLA-DQB1 SSO typing kit showed good performance for intermediate-resolution HLA-DQB1 typing in clinical laboratory for organ transplantation in Koreans.
DNA-based HLA typing is an essential step in the organ and hematopoietic stem cell transplantations. Intermediate-resolution HLA-DQ typing has gained importance in organ transplantation as HLA-DQ antibody is the most frequent de novo donor-specific HLA antibody (DSA) associated with poor transplantation outcome, and epitope analysis of HLA-DQ antigens is critical in the identification of DS [1]. HLA-DQ typing based on the reverse sequence-specific oligonucleotide (SSO) probes method using Luminex (Luminex Corp., Austin, TX, USA), which provides intermediate-resolution DNA typing, is being used increasingly [2, 3]. Although the performance of HLA-DRB1 PCR-SSO kit using Luminex was recently reported [4], there is no report on HLA-DQB1 PCR-SSO kit performance using Luminex. Therefore, we evaluated the performance of LIFECODES HLA-DQB1 SSO typing kit using Luminex in Korean individuals.
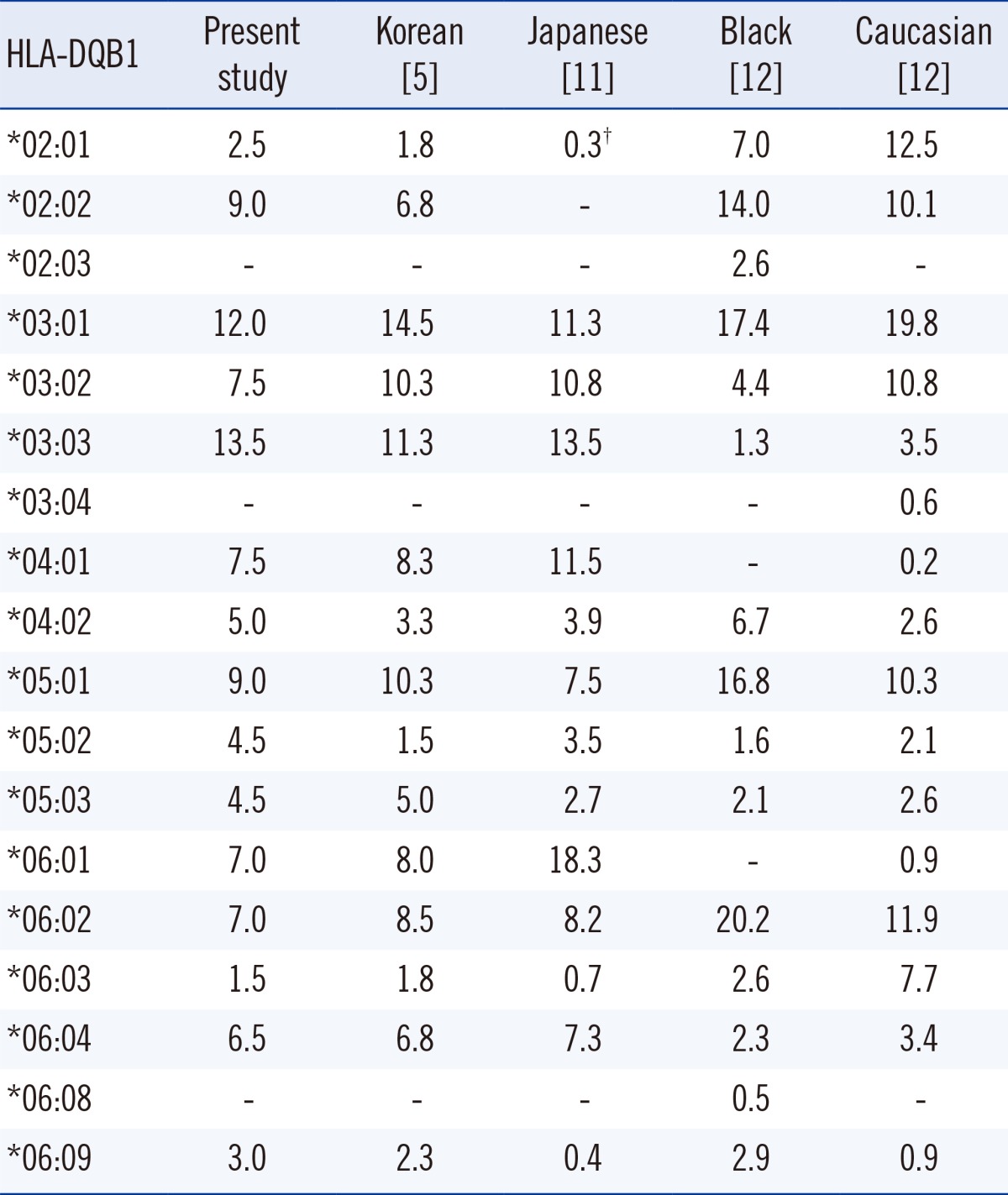
Blood samples were collected from 100 individuals referred to the Seoul National University Hospital between July 2013 and January 2014 for high-resolution HLA-DQB1 typing. High-resolution HLA-DQB1 typing was performed by using the AlleleSEQR HLA-DQB1 sequence-based typing (SBT) kit (Abbott Molecular, Abbott Park, IL, USA). The HLA frequencies of our patients were not different from those previously reported in Koreans (Table 1) [5, 6]. All HLA-DQB1 alleles previously reported with a frequency of ≥0.1% in Koreans were included in our study with more than 3 subjects for each allele. This study was approved by the institutional review board of Seoul National University Hospital (1401-024-547).
Genomic DNA was extracted from the peripheral blood samples by using the QuickGene-Mini80 DNA Isolation System (Fujifilm, Tokyo, Japan) and preserved at -70℃. HLA-DQB1 typing was performed by using the LIFECODES HLA-DQB1 SSO typing kit (Immucor, Stamford, CT, USA). PCR mixture was prepared with 15 µL of the LIFECODES Master Mix (Immucor), 200 ng of genomic DNA, and 2.5 U Taq polymerase in a final volume of 50 µL and then treated with the following: denaturation at 95℃ for 5 min; 40 cycles of amplification (8 cycles: 95℃ for 30 sec, 60℃ for 45 sec, 72℃ for 45 sec, and 32 cycles: 95℃ for 30 sec, 63℃ for 45 sec, 72℃ for 45 sec); and extension at 72℃ for 15 min. Hybridization was performed under the following conditions: 97℃ for 5 min, 47℃ for 30 min, and 56℃ for 10 min with 15 µL probe mix and 5 µL of the PCR product. The samples were diluted with 170 µL of the 1:200 pre-diluted streptavidin-phycoerythrin solution and analyzed within 30 min by using the Luminex 200 system (Luminex Corp.). Lot-specific background control value was subtracted from the raw median fluorescence intensity (MFI) value of the sample to produce the background-corrected data. The background-corrected data were divided by the background- corrected values for the corresponding consensus probe producing the normalized data (adjusted MFI value). The probe-hit pattern was compared with the common and well-documented (CWD) HLA alleles Probe Hit Tables (IMGT/HLA Sequence Database Release 3.11.0) by using the MatchIT DNA program (Immucor).
HLA-DQB1 SBT was performed by using the AlleleSEQR HLA-DQB1 SBT kit (Abbott Molecular, USA). PCR mixture was prepared with 8 µL of PCR Master Mix, 40-80 ng of genomic DNA, and 0.1 µL of AmpliTaq Gold polymerase (Abbott Molecular, USA) in a final volume of 10 µL. PCR was performed under the following conditions: denaturation at 95℃ for 10 min and 36 cycles of amplification (96℃ for 20 sec, 60℃ for 30 sec, 72℃ for 3 min). ExoSAP-IT (3 µL) was added and incubated at 37℃ for 15-30 min and then at 80℃ for 15 min. The PCR product was diluted with Tris-EDTA buffer in a 1:2 (v/v) ratio. The sequencing mix (8 µL) was added with 25 thermal cycles (96℃ for 20 sec, 50℃ for 30 sec, and 60℃ for 2 min). Sodium acetate/ EDTA buffer (2 µL) and absolute ethanol (25 µL) were added. After vigorous vortexing and centrifugation at 2,000 g for 30 min, the supernatant was removed. The step of adding of 50 µL of 80% ethanol and centrifugation at 2,000 g for 5 min were repeated twice. Fifteen microliter of 0.3 mM EDTA was added and loaded onto the ABI 3730XL DNA Analyzer (Applied Biosystems). The sequence data were processed by using the Assign SBT Program (Conexio Genomics, Fremantle Western Australia, Australia).
We assigned HLA-DQB1 alleles to a 4-digit level by using the LIFECODES HLA-DQB1 SSO typing kit and compared them with the high-resolution results obtained from SBT by using the AlleleSEQR HLA-DQB1 kit. The results of analysis of HLA-DQB1 allelic combinations are shown in Table 2. No sample showed ambiguity in the assignment of 4-digit HLA-DQB1 allele with the LIFECODES HLA-DQB1 SSO typing kit; the results were in complete concordance with those of high-resolution typing from AlleleSEQR HLA-DQB1 SBT to the 4-digit level.
Among the 100 sample analyzed, three samples required adjustment of false reactions (3/100, 3.0%). Two samples with DQB1*03:03/*06:01 showed false-positive results in probe 253, and 1 sample with DQB1*04:02/*05:02 showed false-negative result in probe 217. We tested an additional sample with DQB1 *03:03/*06:01 and 2 samples with DQB1*04:02/*05:02 to check if these false reactions are repetitive. One sample with DQB1*03:03/*06:01 showed the same false-positive reaction in probe 253, but 2 samples with DQB1*04:02/*05:02 did not show false reaction. The MFI values (raw/adjusted) of probe 253 for the 3 samples with DQB1*03:03/*06:01 were 1,304/0.433, 1,111/0.354, and 668/0.248. The cut-off values of the adjusted MFI value for positive and negative assignments for probe 253 were 0.199 and 0.163. The MFI value (raw/adjusted) of probe 217 for 1 sample with DQB1*04:02/*05:02 was 732/0.230. The cut-off values for positive and negative assignment for probe 217 were 0.374 and 0.458. Those false reactions did not result in ambiguity or change in the HLA allele assignment by MatchIT DNA program, but 'Review Probes' for probe 253 or 217 was recommended by MatchIT DNA program for the suggested assignment of DQB1*03:03/*06:01 or DQB1*04:02/*05:02.
Intermediate-resolution HLA-DQB1 typing is not routinely performed in organ transplantation. However, recently, the accumulating evidences suggested the role of HLA-DQB1 allele and HLA antibodies reactive to donor-specific HLA-DQB1 alleles [7, 8, 9]. In Eurotransplant, some centers provide HLA-DQB1 results for virtual cross-matching [10]. Bead-based HLA test developed for intermediate-resolution HLA typing using Luminex has also been introduced and is being increasingly used with some advantages such as simple procedure, high-throughput, and low cost compared with sequence-based HLA typing.
We could assign HLA-DQB1 alleles to 4 digit-level without ambiguity using Luminex SSO test by MatchIT DNA program based on the CWD HLA alleles, and the results were 100% concordant with those of SBT, which was slightly different from the 20.4% ambiguity previously reported by using the HLA-DRB1 SSO kit [4]. It is probably owing to lesser allelic polymorphism of HLA-DQB1 locus and the difference in the database used for allelic assignment. In our study, 3 out of 100 samples (3.0%) required adjustment for false reactions. One sample with DQB1*04:02/*05:02 showed false-negative result in probe 217, and 2 samples with DQB1*03:03/*06:01 showed false-positive result in probe 253. The frequency of false reactions in the HLA-DQB1 HLA SSO kit was lower than that in the HLA-DRB1 HLA SSO kit (21.2%) [4], probably owing to the manufacturer difference of the HLA SSO kit or in the property of amplification of HLA-DRB1 and DQB1 loci; further study is needed to clarify the cause for the difference in the frequency values between the kits. Although false-negative result in probe 217 was not repetitive in 2 additional samples with DQB1*04:02/*05:02, false-positive result in probe 253 in samples with DQB1*03:03/*06:01 was consistent. The expected phenotype frequency of DQB1*03:03/*06:01 combination in Koreans is 1.89%, which is higher than that of Caucasians (0.06%), as inferred from HLA-DQB1 allele frequencies (Table 1). The cause of false-positive result for probe 253, especially in samples with DQB1*03:03/*06:01 allelic combination, should be further clarified and improved in the near future. However, these false reactions did not result in ambiguity or change in the HLA allele assignment, because they could be easily detected by the interpretation program.
Although the sample size was small in this study, we included all the DQB1 alleles with a gene frequency of ≥ 0.01% in Koreans with more than 3 subjects for each allele. We could assign HLA-DQB1 alleles up to 4 digit-level without ambiguity and with 100% concordance with the SBT results. In conclusion, LIFECODES HLA-DQB1 SSO typing kit demonstrated good performance for intermediate-resolution HLA-DQB1 typing and may have clinical applications during organ transplantation in Korean patients.
References
1. Tambur AR, Rosati J, Roitberg S, Glotz D, Friedewald JJ, Leventhal JR. Epitope analysis of HLA-DQ antigens: what does the antibody see? Transplantation. 2014; 98:157–166. PMID: 25003284.
2. Lachmann N, Todorova K, Schulze H, Schönemann C. Luminex® and its applications for solid organ transplantation, hematopoietic stem cell transplantation, and transfusion. Transfus Med Hemother. 2013; 40:182–189. PMID: 23922543.

3. Trajanoski D, Fidler SJ. HLA typing using bead-based methods. Methods Mol Biol. 2012; 882:47–65. PMID: 22665228.

4. Testi M, Iannelli S, Testa G, Troiano M, Capelli S, Fruet F, et al. Evaluation of DRB1 high resolution typing by a new SSO-based Luminex method. Mol Biol Rep. 2012; 39:13–16. PMID: 21424786.

5. Song EY, Park MH, Kang SJ, Park HJ, Kim BC, Tokunaga K, et al. HLA class II allele and haplotype frequencies in Koreans based on 107 families. Tissue Antigens. 2002; 59:475–486. PMID: 12445317.

6. Roh EY, Park MH, Park H, Park DH, Choi JB, Kim SJ, et al. Association of HLA-DR and -DQ genes with narcolepsy in Koreans: comparison with two control groups, randomly selected subjects and DRB1*1501-DQB1*0602--positive subjects. Hum Immunol. 2006; 67:749–755. PMID: 17002906.
7. DeVos JM, Gaber AO, Knight RJ, Land GA, Suki WN, Gaber LW, et al. Donor-specific HLA-DQ antibodies may contribute to poor graft outcome after renal transplantation. Kidney Int. 2012; 82:598–604. PMID: 22622504.

8. Willicombe M, Brookes P, Sergeant R, Santos-Nunez E, Steggar C, Galliford J, et al. De novo DQ donor-specific antibodies are associated with a significant risk of antibody-mediated rejection and transplant glomerulopathy. Transplantation. 2012; 94:172–177. PMID: 22735711.

9. Tambur AR, Leventhal JR, Friedewald JJ, Ramon DS. The complexity of human leukocyte antigen (HLA)-DQ antibodies and its effect on virtual crossmatching. Transplantation. 2010; 90:1117–1124. PMID: 20847715.

10. Claas FH, Rahmel A, Doxiadis II. Enhanced kidney allocation to highly sensitized patients by the acceptable mismatch program. Transplantation. 2009; 88:447–452. PMID: 19696624.

11. Saito S, Ota S, Yamada E, Inoki H, Ota M. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens. 2000; 56:522–529. PMID: 11169242.

12. Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, Rybicki BA, et al. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet. 2003; 73:720–735. PMID: 14508706.
Table 2
HLA-DQB1 assignment by using the LIFECODE HLA-DQB1 SSO typing kit in 100 Koreans
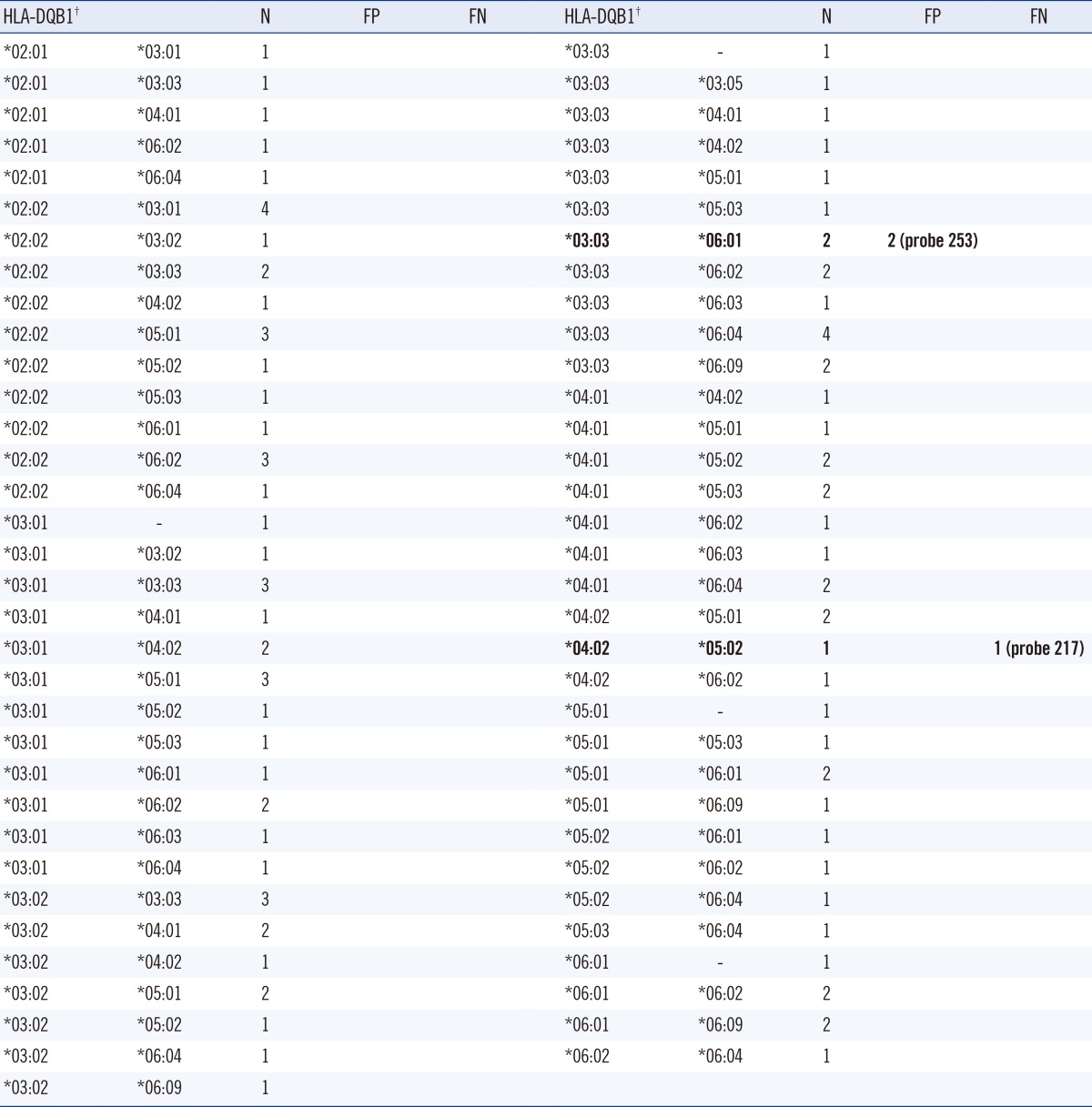
The assigned SSO results were 100% concordant with the AlleleSEQR HLA-DQB1 sequence-based typing (SBT) results.
†High resolution HLA-DQB1 typing was performed by using the AlleleSEQR HLA-DQB1 SBT kit. HLA-DQB1 allelic combinations with false-positive or false-negative probe by using the LIFECODE HLA-DQB1 SSO typing kit are shown in bold font.
Abbreviations: SSO, sequence-specific oligonucleotide; N, number; FP, false positive; FN, false negative.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download